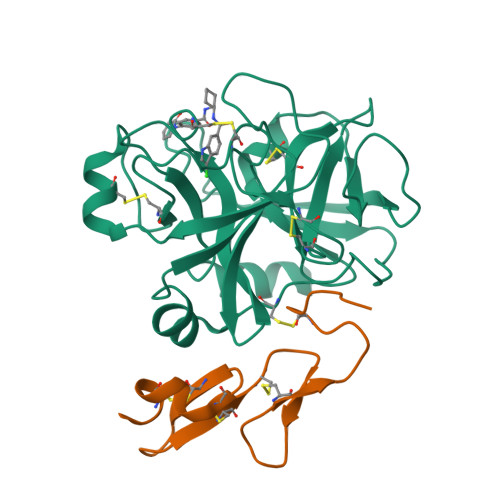
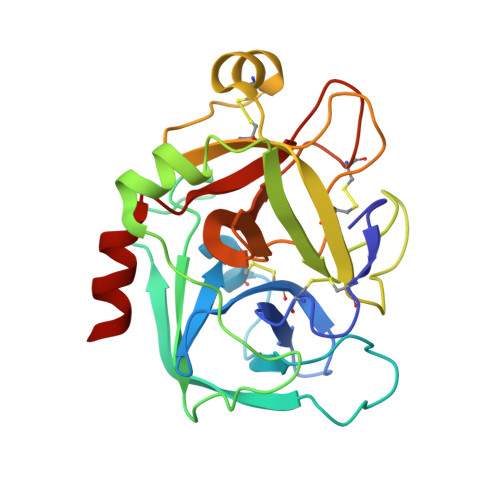
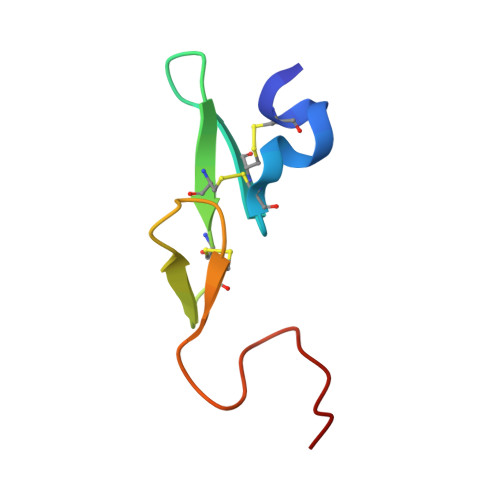
SAR and X-ray structures of enantiopure 1,2-cis-(1R,2S)-cyclopentyldiamine and cyclohexyldiamine derivatives as inhibitors of coagulation Factor Xa
Qiao, J.X., Chang, C.-H., Cheney, D.L., Morin, P.E., Wang, G.Z., King, S.R., Wang, T.C., Rendina, A.R., Luettgen, J.M., Knabb, R.M., Wexler, R.R., Lam, P.Y.S.(2007) Bioorg Med Chem Lett 17: 4419-4427
- PubMed: 17588746
- DOI: https://doi.org/10.1016/j.bmcl.2007.06.029
- Primary Citation of Related Structures:
2P93, 2P94, 2P95 - PubMed Abstract:
In the search of Factor Xa (FXa) inhibitors structurally different from the pyrazole-based series, we identified a viable series of enantiopure cis-(1R,2S)-cycloalkyldiamine derivatives as potent and selective inhibitors of FXa. Among them, cyclohexyldiamide 7 and cyclopentyldiamide 9 were the most potent neutral compounds, and had good anticoagulant activity comparable to the pyrazole-based analogs. Crystal structures of 7-FXa and 9-FXa illustrate binding similarities and differences between the five- and the six-membered core systems, and provide rationales for the observed SAR of P1 and linker moieties.
Organizational Affiliation:
Bristol-Myers Squibb Company, Research and Development, PO Box 5400, Princeton, NJ 08543-5400, USA. jennifer.qiao@bms.com