A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Torelli, A.T., Krucinska, J., Wedekind, J.E.(2007) RNA 13: 1052-1070
- PubMed: 17488874
- DOI: https://doi.org/10.1261/rna.510807
- Primary Citation of Related Structures:
2P7D, 2P7E, 2P7F - PubMed Abstract:
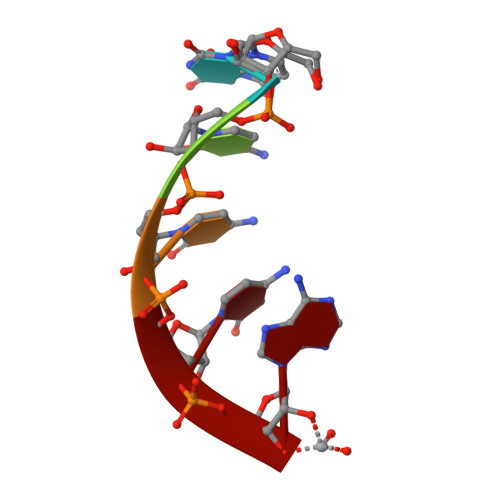
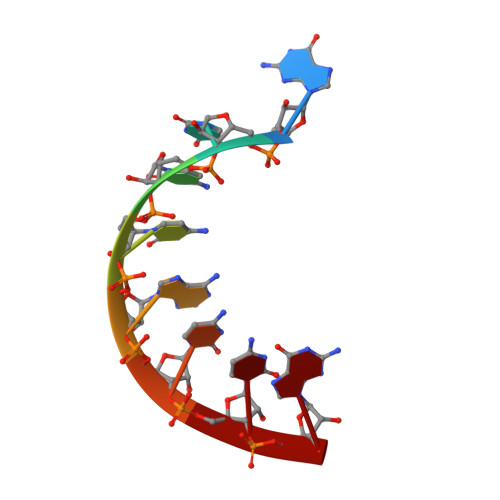
The potential for water to participate in RNA catalyzed reactions has been the topic of several recent studies. Here, we report crystals of a minimal, hinged hairpin ribozyme in complex with the transition-state analog vanadate at 2.05 A resolution. Waters are present in the active site and are discussed in light of existing views of catalytic strategies employed by the hairpin ribozyme. A second structure harboring a 2',5'-phosphodiester linkage at the site of cleavage was also solved at 2.35 A resolution and corroborates the assignment of active site waters in the structure containing vanadate. A comparison of the two structures reveals that the 2',5' structure adopts a conformation that resembles the reaction intermediate in terms of (1) the positioning of its nonbridging oxygens and (2) the covalent attachment of the 2'-O nucleophile with the scissile G+1 phosphorus. The 2',5'-linked structure was then overlaid with scissile bonds of other small ribozymes including the glmS metabolite-sensing riboswitch and the hammerhead ribozyme, and suggests the potential of the 2',5' linkage to elicit a reaction-intermediate conformation without the need to form metalloenzyme complexes. The hairpin ribozyme structures presented here also suggest how water molecules bound at each of the nonbridging oxygens of G+1 may electrostatically stabilize the transition state in a manner that supplements nucleobase functional groups. Such coordination has not been reported for small ribozymes, but is consistent with the structures of protein enzymes. Overall, this work establishes significant parallels between the RNA and protein enzyme worlds.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Rochester, NY 14642, USA.