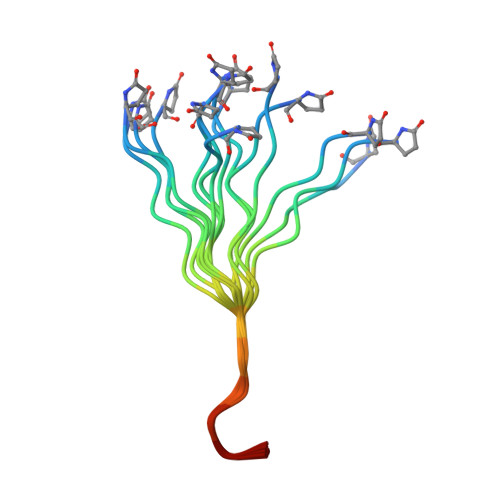
NMR Solution Structure of Neurotensin in Membrane-Mimetic Environments: Molecular Basis for Neurotensin Receptor Recognition.
Coutant, J., Curmi, P.A., Toma, F., Monti, J.P.(2007) Biochemistry 46: 5656-5663
- PubMed: 17441729
- DOI: https://doi.org/10.1021/bi602567p
- Primary Citation of Related Structures:
2OYV, 2OYW - PubMed Abstract:
Neurotensin (NT) is a 13-residue neuropeptide that exerts multiple biological functions in the central and peripheral nervous system. Little is known about the structure of this neuropeptide, and what is known only concerns its C-terminal part. We determined here for the first time the structure of the full-length NT in membrane-mimicking environments by means of classical proton-proton distance constraints derived from solution-state NMR spectroscopy. NT was found to have a structure at both its N and C termini, whereas the central region of NT remains highly flexible. In TFE and HFIP solutions, the NT C-terminus presents an extended slightly incurved structure, whereas in DPC it has a beta turn. The N-terminal region of NT possesses great adaptability and accessibility to the microenvironment in the three media studied. Altogether, our work demonstrates a structure of NT fully compatible with its NTR-bound state.
Organizational Affiliation:
Laboratoire de Physique et Biophysique GESVAB EA 3675, Institut des Sciences de la Vigne et du Vin, Université de Bordeaux 2, 146 rue Léo Saignat, 33076 Bordeaux Cedex, France.