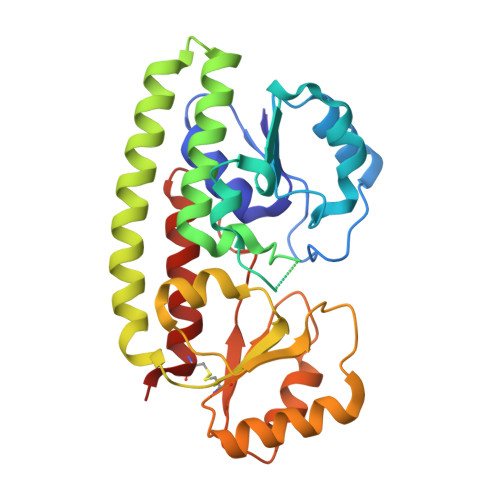
Crystal Structure of the Zinc-binding Transport Protein ZnuA from Escherichia coli Reveals an Unexpected Variation in Metal Coordination.
Li, H., Jogl, G.(2007) J Mol Biology 368: 1358-1366
- PubMed: 17399739
- DOI: https://doi.org/10.1016/j.jmb.2007.02.107
- Primary Citation of Related Structures:
2OSV - PubMed Abstract:
Bacterial ATP-binding cassette transport systems for high-affinity uptake of zinc and manganese use a cluster 9 solute-binding protein. Structures of four cluster 9 transport proteins have been determined previously. However, the structural determinants for discrimination between zinc and manganese remain under discussion. To further investigate the variability of metal binding sites in bacterial transporters, we have determined the structure of the zinc-bound transport protein ZnuA from Escherichia coli to 1.75 A resolution. The overall structure of ZnuA is similar to other solute-binding transporters. A scaffolding alpha-helix forms the backbone for two structurally related globular domains. The metal-binding site is located at the domain interface. The bound zinc ion is coordinated by three histidine residues (His78, His161 and His225) and one glutamate residue (Glu77). The functional role of Glu77 for metal binding is unexpected, because this residue is not conserved in previously determined structures of zinc and manganese-specific transport proteins. The observed metal coordination by four protein residues differs significantly from the zinc-binding site in the ZnuA transporter from Synechocystis 6803, which binds zinc via three histidine residues. In addition, the E. coli ZnuA structure reveals the presence of a disulfide bond in the C-terminal globular domain that is not present in previously determined cluster 9 transport protein structures.
- Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA.
Organizational Affiliation: