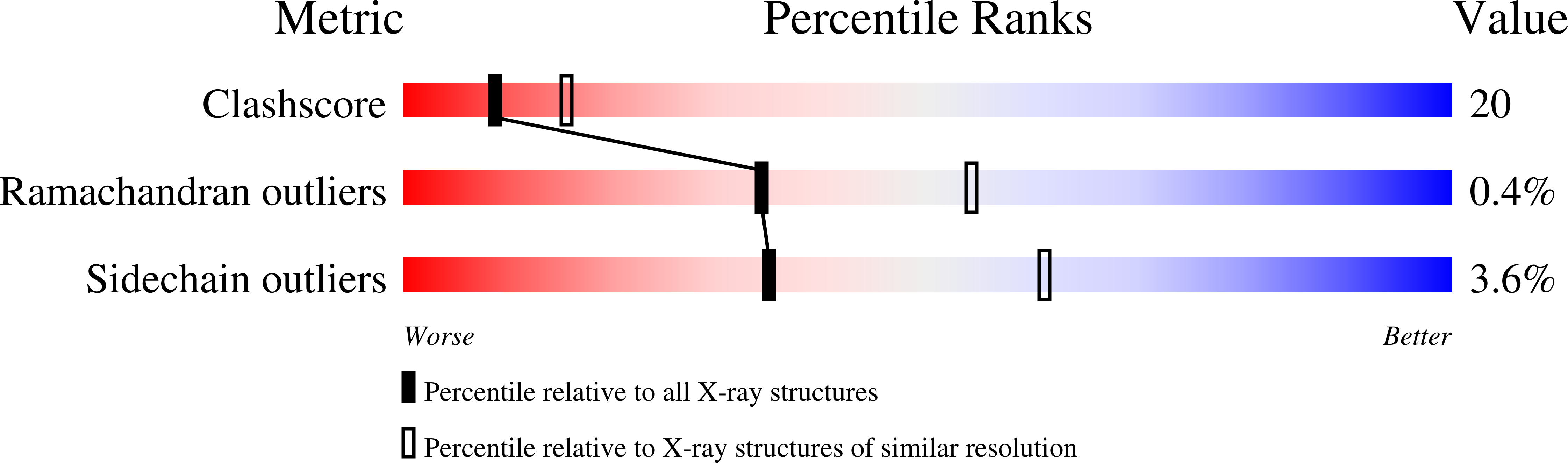Structural basis for glutamate racemase inhibition
Kim, K.H., Bong, Y.J., Park, J.K., Shin, K.J., Hwang, K.Y., Kim, E.E.(2007) J Mol Biology 372: 434-443
- PubMed: 17658548
- DOI: https://doi.org/10.1016/j.jmb.2007.05.003
- Primary Citation of Related Structures:
2OHG, 2OHO, 2OHV - PubMed Abstract:
D-Glutamic acid is a required biosynthetic building block for peptidoglycan, and the enzyme glutamate racemase (GluR) catalyzes the inter-conversion of D and L-glutamate enantiomers. Therefore, GluR is considered as an attractive target for the design of new antibacterial drugs. Here, we report the crystal structures of GluR from Streptococcus pyogenes in both inhibitor-free and inhibitor-bound forms. The inhibitor free GluR crystallized in two different forms, which diffracted to 2.25 A and 2.5 A resolution, while the inhibitor-bound crystal diffracted to 2.5 A resolution. GluR is composed of two domains of alpha/beta protein that are related by pseudo-2-fold symmetry and the active site is located at the domain interface. The inhibitor, gamma-2-naphthylmethyl-D-glutamate, which was reported earlier as a novel potent competitive inhibitor, makes several hydrogen bonds with protein atoms, and the naphthyl moiety is located in the hydrophobic pocket. The inhibitor binding induces a disorder in one of the loops near the active site. In both crystal forms, GluR exists as a dimer and the interactions seen at the dimer interface are almost identical. This agrees well with the results from gel filtration and dynamic light-scattering studies.
Organizational Affiliation:
Life Sciences Division, Korea Institute of Science and Technology, 39-1 Hawolkok-Dong, Sungbuk-Gu, Seoul, Korea.