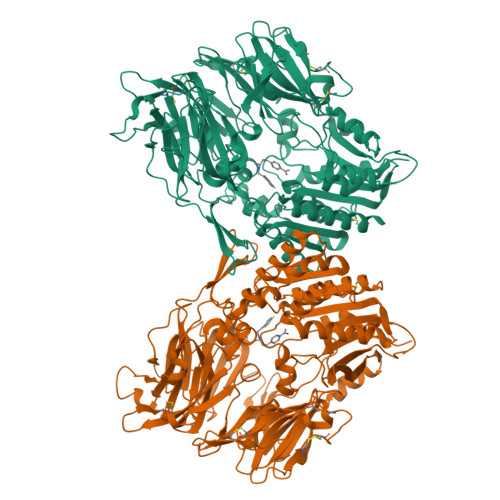
Discovery of non-covalent dipeptidyl peptidase IV inhibitors which induce a conformational change in the active site.
Sheehan, S.M., Mest, H.J., Watson, B.M., Klimkowski, V.J., Timm, D.E., Cauvin, A., Parsons, S.H., Shi, Q., Canada, E.J., Wiley, M.R., Ruehter, G., Evers, B., Petersen, S., Blaszczak, L.C., Pulley, S.R., Margolis, B.J., Wishart, G.N., Renson, B., Hankotius, D., Mohr, M., Zechel, J.C., Michael Kalbfleisch, J., Dingess-Hammond, E.A., Boelke, A., Weichert, A.G.(2007) Bioorg Med Chem Lett 17: 1765-1768
- PubMed: 17239592
- DOI: https://doi.org/10.1016/j.bmcl.2006.12.074
- Primary Citation of Related Structures:
2OGZ - PubMed Abstract:
A series of non-covalent inhibitors of the serine protease dipeptidyl peptidase IV (DPP-IV) were found to adopt a U-shaped binding conformation in X-ray co-crystallization studies. Remarkably, Tyr547 undergoes a 70 degrees side-chain rotation to accommodate the inhibitor and allows access to a previously unexposed area of the protein backbone for hydrogen bonding.
Organizational Affiliation:
Lilly Research Laboratories, A Division of Eli Lilly and Company, Indianapolis, IN 46285, USA. SHEEHAN_SCOTT@LILLY.COM