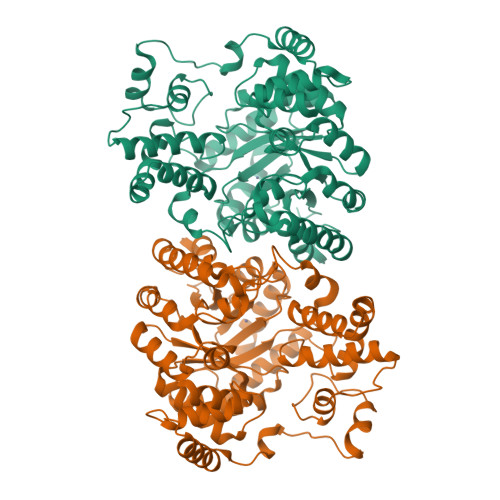
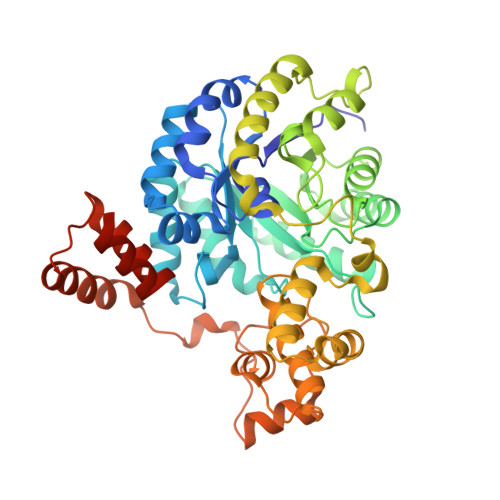
Crystal Structure of the Carboxyltransferase Domain of the Oxaloacetate Decarboxylase Na(+) Pump from Vibrio cholerae.
Studer, R., Dahinden, P., Wang, W.W., Auchli, Y., Li, X.D., Dimroth, P.(2007) J Mol Biology 367: 547-557
- PubMed: 17270211
- DOI: https://doi.org/10.1016/j.jmb.2006.12.035
- Primary Citation of Related Structures:
2NX9 - PubMed Abstract:
Oxaloacetate decarboxylase is a membrane-bound multiprotein complex that couples oxaloacetate decarboxylation to sodium ion transport across the membrane. The initial reaction catalyzed by this enzyme machinery is the carboxyl transfer from oxaloacetate to the prosthetic biotin group. The crystal structure of the carboxyltransferase at 1.7 A resolution shows a dimer of alpha(8)beta(8) barrels with an active site metal ion, identified spectroscopically as Zn(2+), at the bottom of a deep cleft. The enzyme is completely inactivated by specific mutagenesis of Asp17, His207 and His209, which serve as ligands for the Zn(2+) metal ion, or by Lys178 near the active site, suggesting that Zn(2+) as well as Lys178 are essential for the catalysis. In the present structure this lysine residue is hydrogen-bonded to Cys148. A potential role of Lys178 as initial acceptor of the carboxyl group from oxaloacetate is discussed.
- Biomolecular Research, Paul Scherrer Institut, CH-5232 Villigen, Switzerland.
Organizational Affiliation: