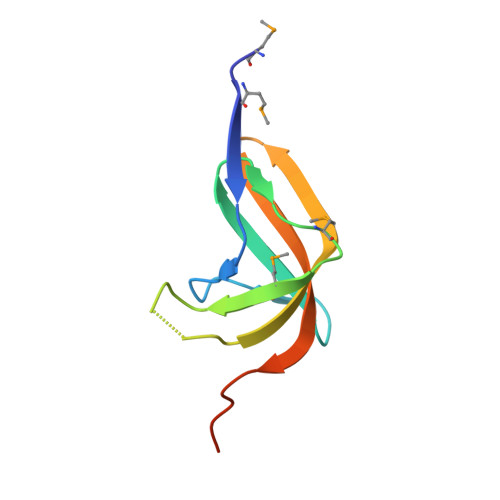
Crystal Structure of the Hypothetical protein YtmB from Bacillus subtilis subsp. (subtilis str. 168), Northeast Structural Genomics target SR466
Zhou, W., Forouhar, F., Seetharaman, J., Wang, D., Cunningham, K., Ma, L.-C., Fang, Y., Xiao, R., Baran, M.C., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.