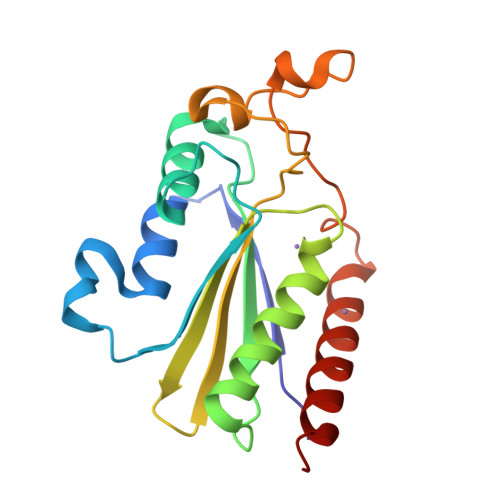
The Structure of the Minimal Relaxase Domain of MobA at 2.1 A Resolution.
Monzingo, A.F., Ozburn, A., Xia, S., Meyer, R.J., Robertus, J.D.(2007) J Mol Biology 366: 165-178
- PubMed: 17157875
- DOI: https://doi.org/10.1016/j.jmb.2006.11.031
- Primary Citation of Related Structures:
2NS6 - PubMed Abstract:
The plasmid R1162 encodes proteins that enable its conjugative mobilization between bacterial cells. It can transfer between many different species and is one of the most promiscuous of the mobilizable plasmids. The plasmid-encoded protein MobA, which has both nicking and priming activities on single-stranded DNA, is essential for mobilization. The nicking, or relaxase, activity has been localized to the 186 residue N-terminal domain, called minMobA. We present here the 2.1 A X-ray structure of minMobA. The fold is similar to that seen for two other relaxases, TraI and TrwC. The similarity in fold, and action, suggests these enzymes are evolutionary homologs, despite the lack of any significant amino acid similarity. MinMobA has a well- defined target DNA called oriT. The active site metal is observed near Tyr25, which is known to form a phosphotyrosine adduct with the substrate. A model of the oriT substrate complexed with minMobA has been made, based on observed substrate binding to TrwC and TraI. The model is consistent with observations of substrate base specificity, and provides a rationalization for elements of the likely enzyme mechanism.
Organizational Affiliation:
Institute of Cellular and Molecular Biology, Department of Chemistry and Biochemistry, 1 University Station A5300, University of Texas, Austin, TX 78712, USA.