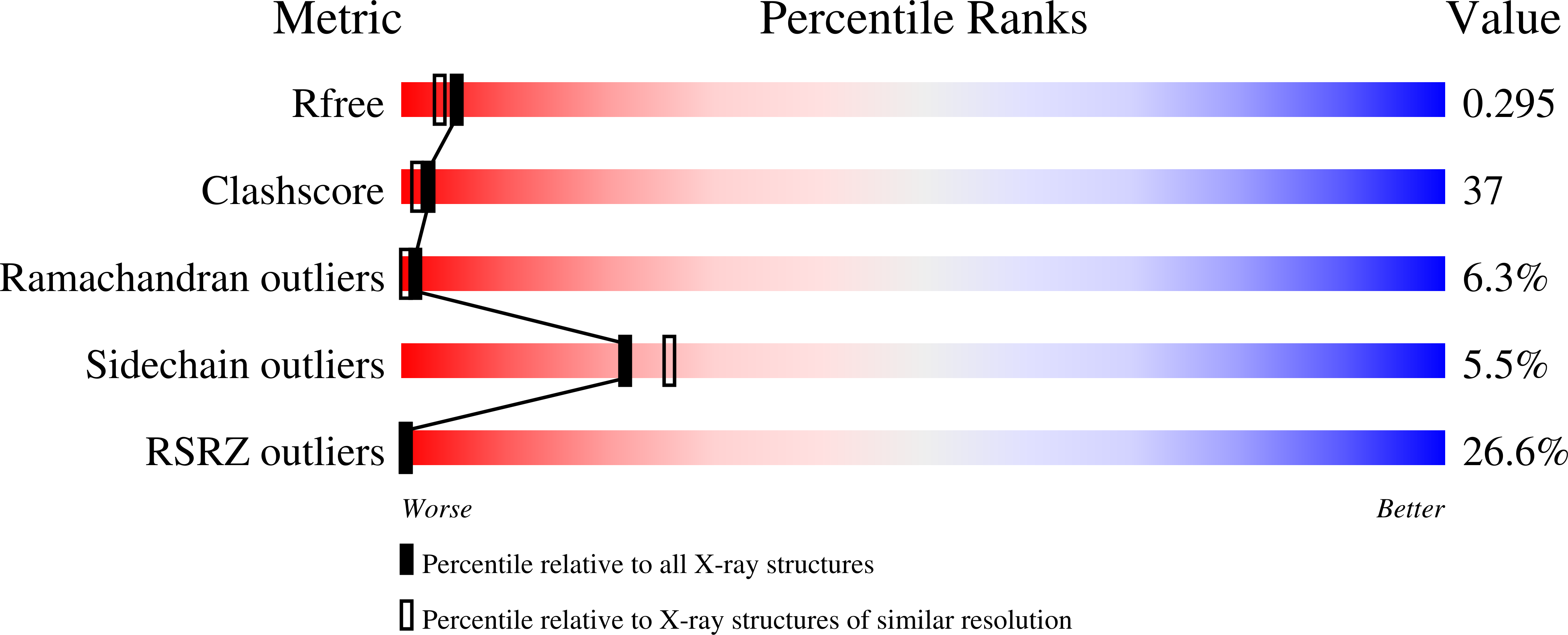
The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis.
Lemieux, M.J., Fischer, S.J., Cherney, M.M., Bateman, K.S., James, M.N.G.(2007) Proc Natl Acad Sci U S A 104: 750-754
- PubMed: 17210913
- DOI: https://doi.org/10.1073/pnas.0609981104
- Primary Citation of Related Structures:
2NR9 - PubMed Abstract:
Rhomboid peptidases are members of a family of regulated intramembrane peptidases that cleave the transmembrane segments of integral membrane proteins. Rhomboid peptidases have been shown to play a major role in developmental processes in Drosophila and in mitochondrial maintenance in yeast. Most recently, the function of rhomboid peptidases has been directly linked to apoptosis. We have solved the structure of the rhomboid peptidase from Haemophilus influenzae (hiGlpG) to 2.2-A resolution. The phasing for the crystals of hiGlpG was provided mainly by molecular replacement, by using the coordinates of the Escherichia coli rhomboid (ecGlpG). The structural results on these rhomboid peptidases have allowed us to speculate on the catalytic mechanism of substrate cleavage in a membranous environment. We have identified the relative disposition of the nucleophilic serine to the general base/acid function of the conserved histidine. Modeling a tetrapeptide substrate in the context of the rhomboid structure reveals an oxyanion hole comprising the side chain of a second conserved histidine and the main-chain NH of the nucleophilic serine residue. In both hiGlpG and ecGlpG structures, a water molecule occupies this oxyanion hole.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, AB, Canada T6G 2H7.