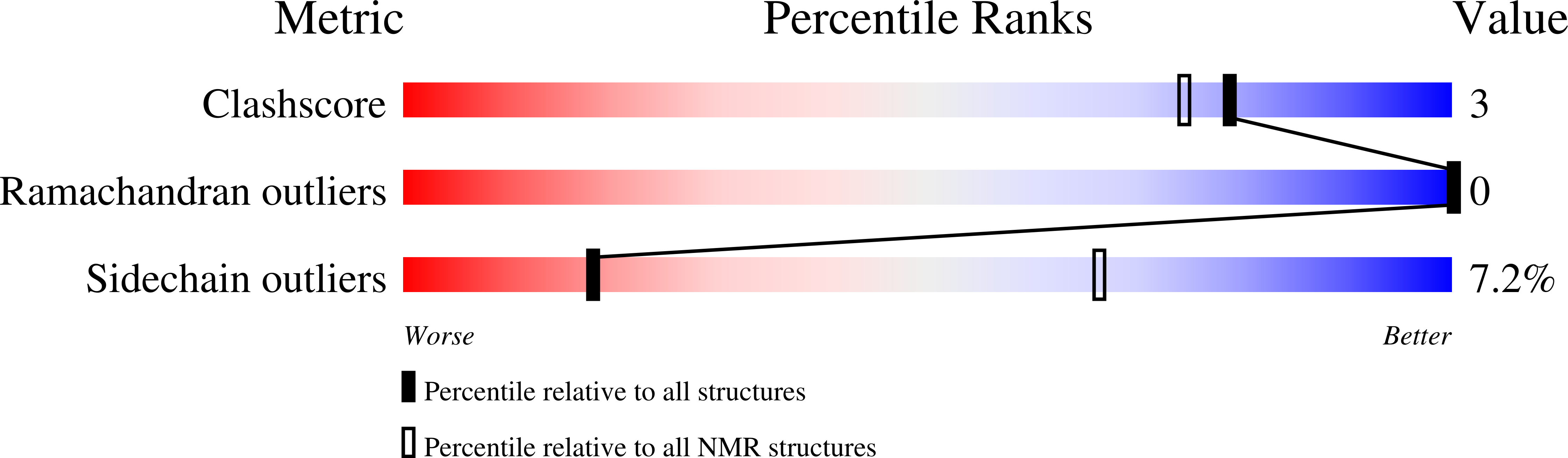Structural and functional insights into the E3 ligase, RNF126.
Krysztofinska, E.M., Martinez-Lumbreras, S., Thapaliya, A., Evans, N.J., High, S., Isaacson, R.L.(2016) Sci Rep 6: 26433-26433
- PubMed: 27193484
- DOI: https://doi.org/10.1038/srep26433
- Primary Citation of Related Structures:
2N9O, 2N9P - PubMed Abstract:
RNF126 is an E3 ubiquitin ligase that collaborates with the BAG6 sortase complex to ubiquitinate hydrophobic substrates in the cytoplasm that are destined for proteasomal recycling. Composed of a trimeric complex of BAG6, TRC35 and UBL4A the BAG6 sortase is also associated with SGTA, a co-chaperone from which it can obtain hydrophobic substrates. Here we solve the solution structure of the RNF126 zinc finger domain in complex with the BAG6 UBL domain. We also characterise an interaction between RNF126 and UBL4A and analyse the competition between SGTA and RNF126 for the N-terminal BAG6 binding site. This work sheds light on the sorting mechanism of the BAG6 complex and its accessory proteins which, together, decide the fate of stray hydrophobic proteins in the aqueous cytoplasm.
Organizational Affiliation:
Department of Chemistry, King's College London, Britannia House, Trinity Street, London, SE1 1DB, UK.