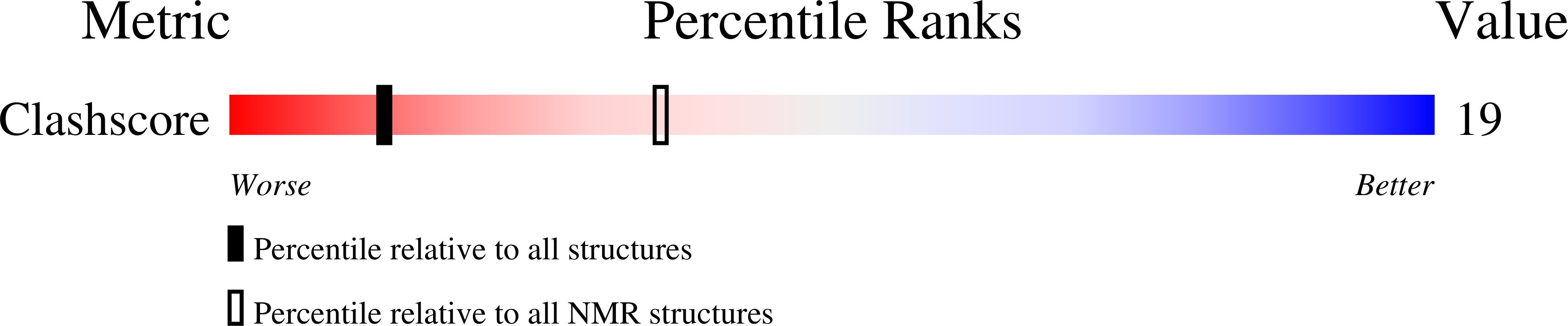
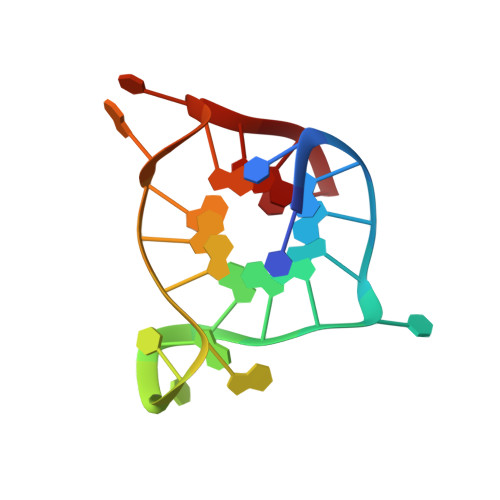
Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome.
De Nicola, B., Lech, C.J., Heddi, B., Regmi, S., Frasson, I., Perrone, R., Richter, S.N., Phan, A.T.(2016) Nucleic Acids Res 44: 6442-6451
- PubMed: 27298260
- DOI: https://doi.org/10.1093/nar/gkw432
- Primary Citation of Related Structures:
2N4Y - PubMed Abstract:
The long terminal repeat (LTR) of the proviral human immunodeficiency virus (HIV)-1 genome is integral to virus transcription and host cell infection. The guanine-rich U3 region within the LTR promoter, previously shown to form G-quadruplex structures, represents an attractive target to inhibit HIV transcription and replication. In this work, we report the structure of a biologically relevant G-quadruplex within the LTR promoter region of HIV-1. The guanine-rich sequence designated LTR-IV forms a well-defined structure in physiological cationic solution. The nuclear magnetic resonance (NMR) structure of this sequence reveals a parallel-stranded G-quadruplex containing a single-nucleotide thymine bulge, which participates in a conserved stacking interaction with a neighboring single-nucleotide adenine loop. Transcription analysis in a HIV-1 replication competent cell indicates that the LTR-IV region may act as a modulator of G-quadruplex formation in the LTR promoter. Consequently, the LTR-IV G-quadruplex structure presented within this work could represent a valuable target for the design of HIV therapeutics.
Organizational Affiliation:
School of Physical and Mathematical Sciences, Nanyang Technological University, Singapore Department of Molecular Medicine, University of Padua, Italy.