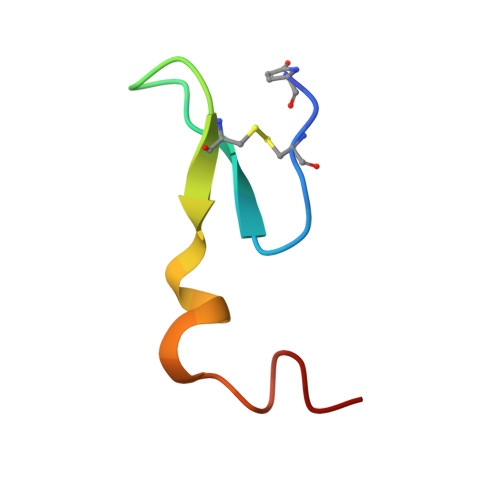
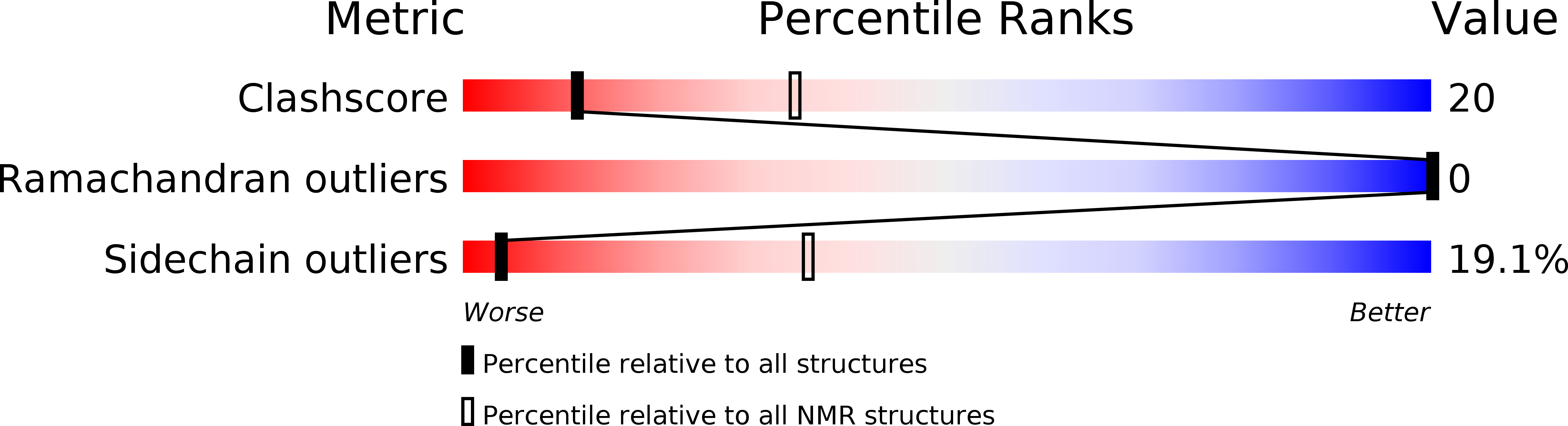A Naturally Occurring Peptide with an Elementary Single Disulfide-Directed beta-Hairpin Fold.
Robinson, S.D., Chhabra, S., Belgi, A., Chittoor, B., Safavi-Hemami, H., Robinson, A.J., Papenfuss, A.T., Purcell, A.W., Norton, R.S.(2016) Structure 24: 293-299
- PubMed: 26774129
- DOI: https://doi.org/10.1016/j.str.2015.11.015
- Primary Citation of Related Structures:
2N24 - PubMed Abstract:
Certain peptide folds, owing to a combination of intrinsic stability and resilience to amino acid substitutions, are particularly effective for the display of diverse functional groups. Such "privileged scaffolds" are valuable as starting points for the engineering of new bioactive molecules. We have identified a precursor peptide expressed in the venom gland of the marine snail Conus victoriae, which appears to belong to a hitherto undescribed class of molluscan neuropeptides. Mass spectrometry matching with the venom confirmed the complete mature peptide sequence as a 31-residue peptide with a single disulfide bond. Solution structure determination revealed a unique peptide fold that we have designated the single disulfide-directed β hairpin (SDH). The SDH fold is highly resistant to thermal denaturation and forms the core of several other multiple disulfide-containing peptide folds, including the inhibitor cystine knot. This elementary fold may offer a valuable starting point for the design and engineering of new bioactive peptides.
Organizational Affiliation:
Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University, 381 Royal Parade, Parkville, VIC 3052, Australia.