Centromeric Alpha-Satellite DNA Adopts Dimeric i-Motif Structures Capped by AT Hoogsteen Base Pairs.
Garavis, M., Escaja, N., Gabelica, V., Villasante, A., Gonzalez, C.(2015) Chemistry 21: 9816-9824
- PubMed: 26013031
- DOI: https://doi.org/10.1002/chem.201500448
- Primary Citation of Related Structures:
2MRZ - PubMed Abstract:
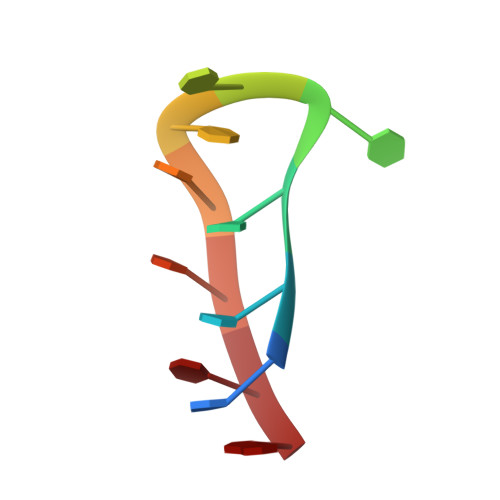
Human centromeric alpha-satellite DNA is composed of tandem arrays of two types of 171 bp monomers; type A and type B. The differences between these types are concentrated in a 17 bp region of the monomer called the A/B box. Here, we have determined the solution structure of the C-rich strand of the two main variants of the human alpha-satellite A box. We show that, under acidic conditions, the C-rich strands of two A boxes self-recognize and form a head-to-tail dimeric i-motif stabilized by four intercalated hemi-protonated C:C(+) base pairs. Interestingly, the stack of C:C(+) base pairs is capped by T:T and Hoogsteen A:T base pairs. The two main variants of the A box adopt a similar three-dimensional structure, although the residues involved in the formation of the i-motif core are different in each case. Together with previous studies showing that the B box (known as the CENP-B box) also forms dimeric i-motif structures, our finding of this non-canonical structure in the A box shows that centromeric alpha satellites in all human chromosomes are able to form i-motifs, which consequently raises the possibility that these structures may play a role in the structural organization of the centromere.
Organizational Affiliation:
Instituto de Química Física Rocasolano, CSIC, Serrano 119, 28006 Madrid (Spain).