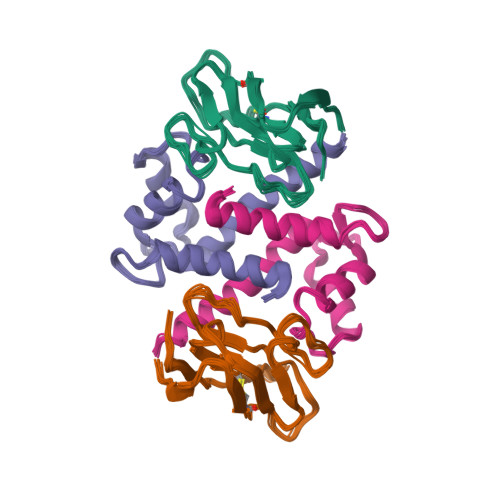
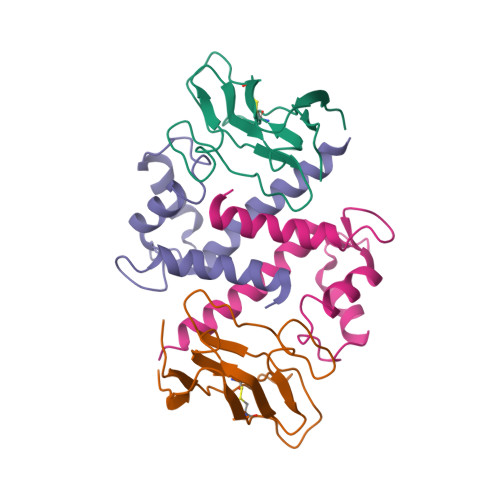
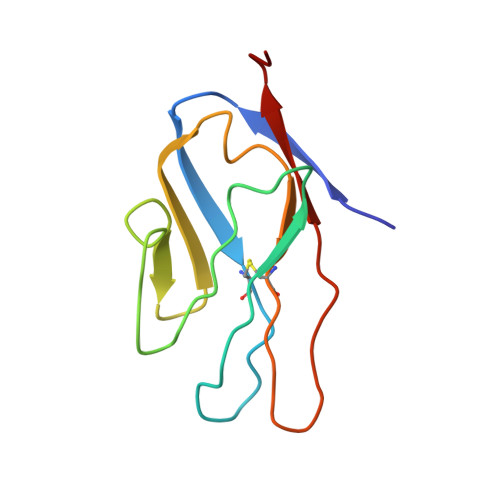
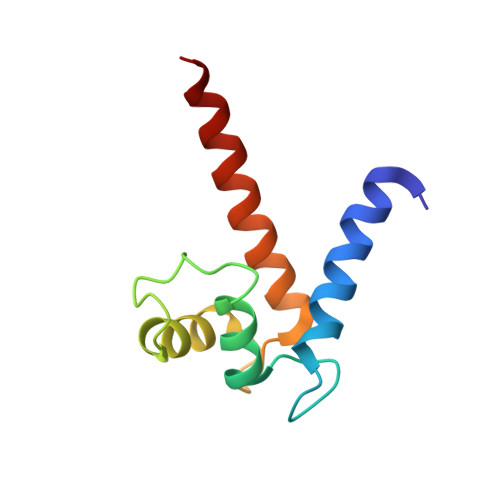
Structural insights into calcium-bound S100P and the V domain of the RAGE complex.
Penumutchu, S.R., Chou, R.H., Yu, C.(2014) PLoS One 9: e103947-e103947
- PubMed: 25084534
- DOI: https://doi.org/10.1371/journal.pone.0103947
- Primary Citation of Related Structures:
2MJW - PubMed Abstract:
The S100P protein is a member of the S100 family of calcium-binding proteins and possesses both intracellular and extracellular functions. Extracellular S100P binds to the cell surface receptor for advanced glycation end products (RAGE) and activates its downstream signaling cascade to meditate tumor growth, drug resistance and metastasis. Preventing the formation of this S100P-RAGE complex is an effective strategy to treat various disease conditions. Despite its importance, the detailed structural characterization of the S100P-RAGE complex has not yet been reported. In this study, we report that S100P preferentially binds to the V domain of RAGE. Furthermore, we characterized the interactions between the RAGE V domain and Ca(2+)-bound S100P using various biophysical techniques, including isothermal titration calorimetry (ITC), fluorescence spectroscopy, multidimensional NMR spectroscopy, functional assays and site-directed mutagenesis. The entropy-driven binding between the V domain of RAGE and Ca(+2)-bound S100P was found to lie in the micromolar range (Kd of ∼ 6 µM). NMR data-driven HADDOCK modeling revealed the putative sites that interact to yield a proposed heterotetrameric model of the S100P-RAGE V domain complex. Our study on the spatial structural information of the proposed protein-protein complex has pharmaceutical relevance and will significantly contribute toward drug development for the prevention of RAGE-related multifarious diseases.
Organizational Affiliation:
Department of Chemistry, National Tsing Hua University, Hsinchu, Taiwan.