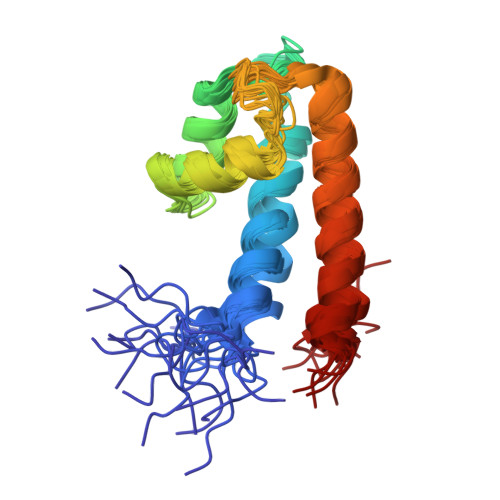
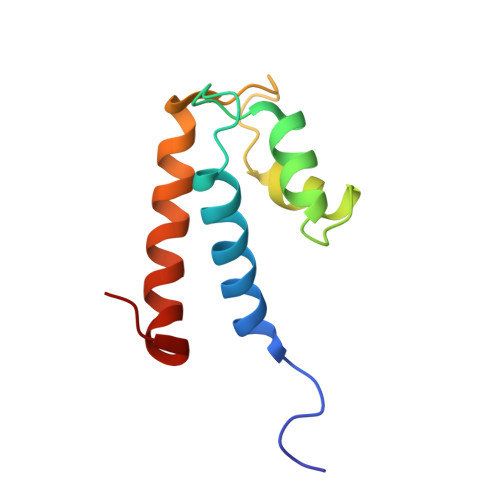
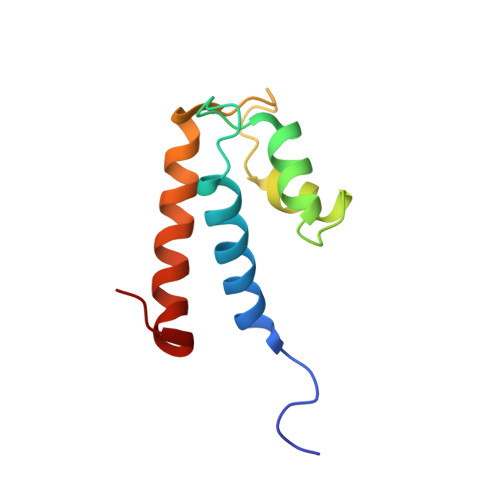
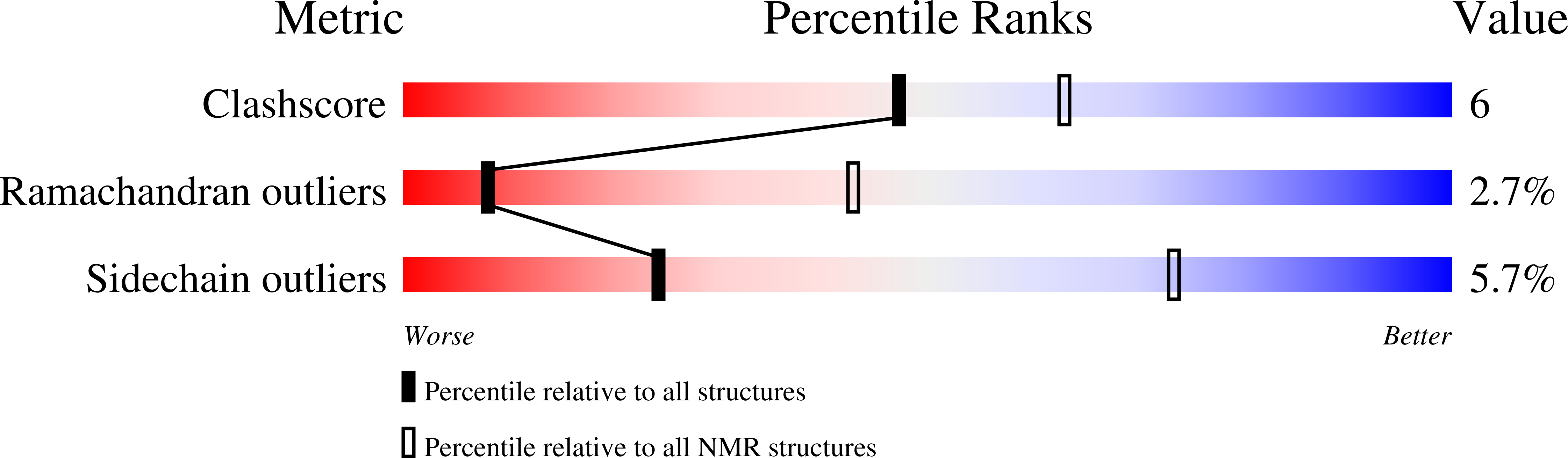The number and location of EF hand motifs dictates the calcium dependence of polycystin-2 function.
Kuo, I.Y., Keeler, C., Corbin, R., Celic, A., Petri, E.T., Hodsdon, M.E., Ehrlich, B.E.(2014) FASEB J 28: 2332-2346
- PubMed: 24558196
- DOI: https://doi.org/10.1096/fj.13-247106
- Primary Citation of Related Structures:
2MHH - PubMed Abstract:
Polycystin 2 (PC2) is a calcium-dependent calcium channel, and mutations to human PC2 (hPC2) are associated with polycystic kidney disease. The C-terminal tail of hPC2 contains 2 EF hand motifs, but only the second binds calcium. Here, we investigate whether these EF hand motifs serve as a calcium sensor responsible for the calcium dependence of PC2 function. Using NMR and bioinformatics, we show that the overall fold is highly conserved, but in evolutionarily earlier species, both EF hands bind calcium. To test whether the EF hand motif is truly a calcium sensor controlling PC2 channel function, we altered the number of calcium binding sites in hPC2. NMR studies confirmed that modified hPC2 binds an additional calcium ion. Single-channel recordings demonstrated a leftward shift in the calcium dependence, and imaging studies in cells showed that calcium transients were enhanced compared with wild-type hPC2. However, biophysics and functional studies showed that the first EF hand can only bind calcium and be functionally active if the second (native) calcium-binding EF hand is intact. These results suggest that the number and location of calcium-binding sites in the EF hand senses the concentration of calcium required for PC2 channel activity and cellular function.
Organizational Affiliation:
2B.E.E., Department of Pharmacology, Yale University School of Medicine, 333 Cedar St, New Haven, CT 06520-8066, USA. barbara.ehrlich@yale.edu.