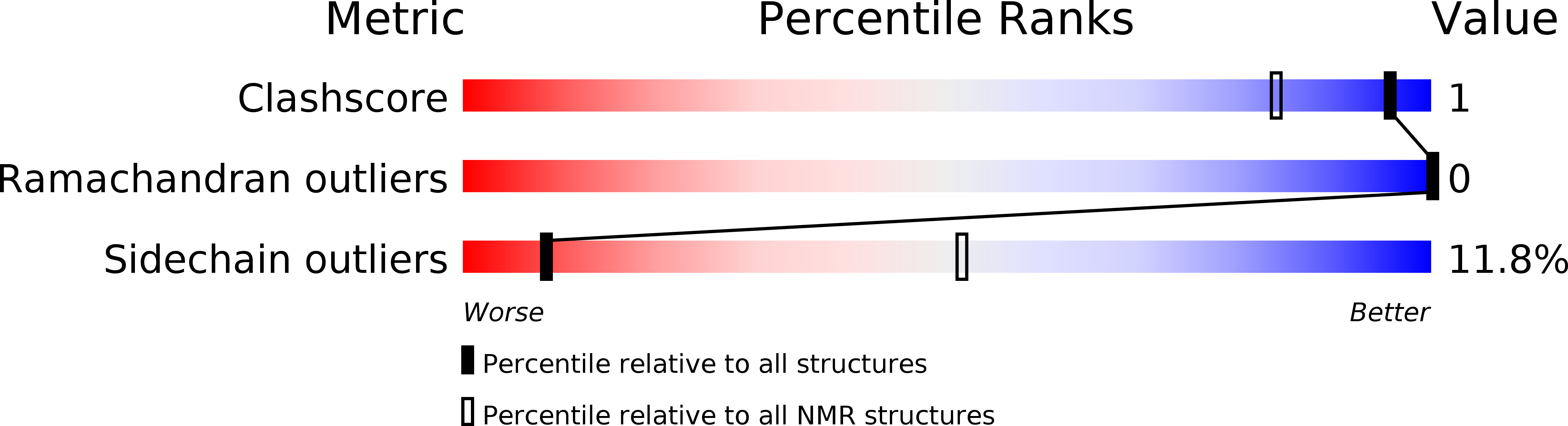Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
Chen, W., Enck, S., Price, J.L., Powers, D.L., Powers, E.T., Wong, C.H., Dyson, H.J., Kelly, J.W.(2013) J Am Chem Soc 135: 9877-9884
- PubMed: 23742246
- DOI: https://doi.org/10.1021/ja4040472
- Primary Citation of Related Structures:
2M9E, 2M9F, 2M9I, 2M9J - PubMed Abstract:
Carbohydrate-aromatic interactions mediate many biological processes. However, the structure-energy relationships underpinning direct carbohydrate-aromatic packing interactions in aqueous solution have been difficult to assess experimentally and remain elusive. Here, we determine the structures and folding energetics of chemically synthesized glycoproteins to quantify the contributions of the hydrophobic effect and CH-π interactions to carbohydrate-aromatic packing interactions in proteins. We find that the hydrophobic effect contributes significantly to protein-carbohydrate interactions. Interactions between carbohydrates and aromatic amino acid side chains, however, are supplemented by CH-π interactions. The strengths of experimentally determined carbohydrate CH-π interactions do not correlate with the electrostatic properties of the involved aromatic residues, suggesting that the electrostatic component of CH-π interactions in aqueous solution is small. Thus, tight binding of carbohydrates and aromatic residues is driven by the hydrophobic effect and CH-π interactions featuring a dominating dispersive component.
Organizational Affiliation:
Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, California 92037, USA.