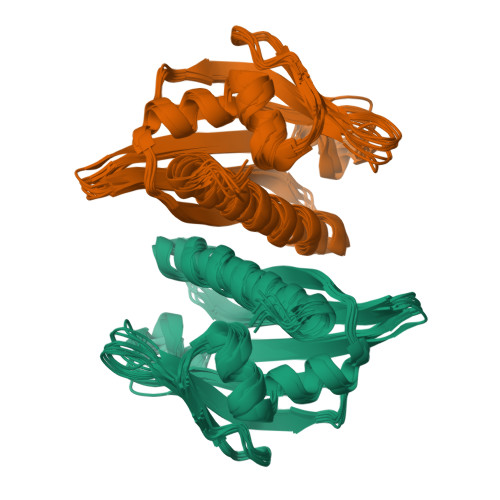
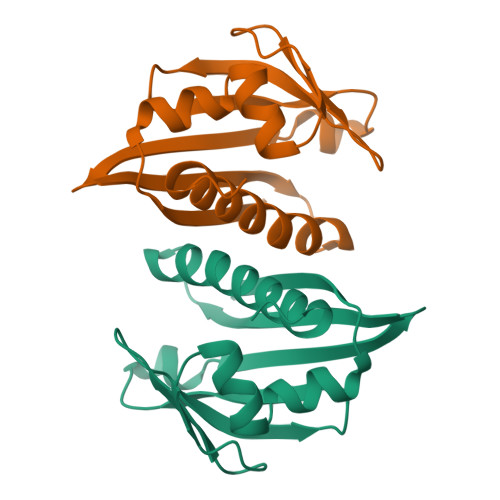
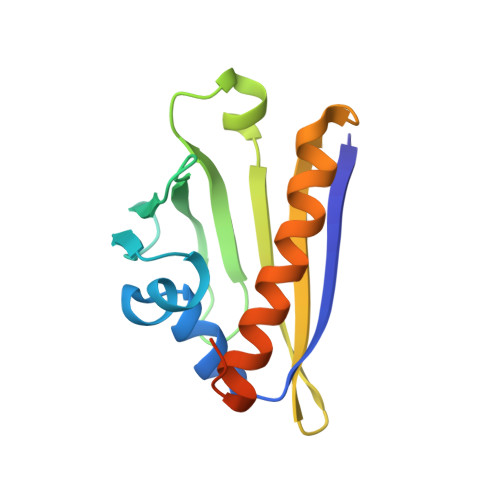
A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Rossi, P., Shi, L., Liu, G., Barbieri, C.M., Lee, H.W., Grant, T.D., Luft, J.R., Xiao, R., Acton, T.B., Snell, E.H., Montelione, G.T., Baker, D., Lange, O.F., Sgourakis, N.G.(2015) Proteins 83: 309-317
- PubMed: 25388768
- DOI: https://doi.org/10.1002/prot.24719
- Primary Citation of Related Structures:
2M89 - PubMed Abstract:
Oligomeric proteins are important targets for structure determination in solution. While in most cases the fold of individual subunits can be determined experimentally, or predicted by homology-based methods, protein-protein interfaces are challenging to determine de novo using conventional NMR structure determination protocols. Here we focus on a member of the bet-V1 superfamily, Aha1 from Colwellia psychrerythraea. This family displays a broad range of crystallographic interfaces none of which can be reconciled with the NMR and SAXS data collected for Aha1. Unlike conventional methods relying on a dense network of experimental restraints, the sparse data are used to limit conformational search during optimization of a physically realistic energy function. This work highlights a new approach for studying minor conformational changes due to structural plasticity within a single dimeric interface in solution.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, Center for Advanced Biotechnology and Medicine, and Northeast Structural Genomics Consortium, Rutgers University, Piscataway, New Jersey, 08854.