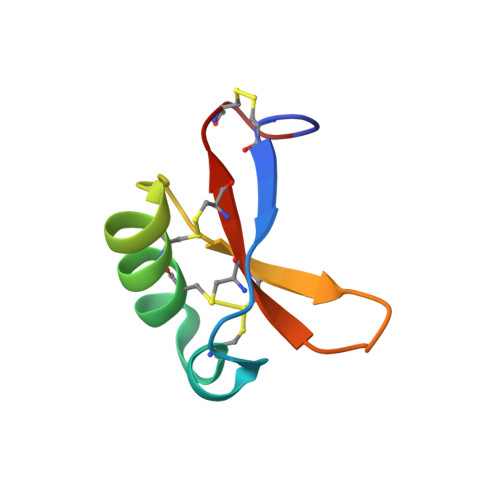
Temperature-dependent conformational change affecting Tyr11 and sweetness loops of brazzein.
Cornilescu, C.C., Cornilescu, G., Rao, H., Porter, S.F., Tonelli, M., Derider, M.L., Markley, J.L., Assadi-Porter, F.M.(2013) Proteins 81: 919-925
- PubMed: 23349025
- DOI: https://doi.org/10.1002/prot.24259
- Primary Citation of Related Structures:
2LY5, 2LY6 - PubMed Abstract:
The sweet protein brazzein, a member of the Csβα fold family, contains four disulfide bonds that lend a high degree of thermal and pH stability to its structure. Nevertheless, a variable temperature study has revealed that the protein undergoes a local, reversible conformational change between 37 and 3°C with a midpoint about 27°C that changes the orientations and side-chain hydrogen bond partners of Tyr8 and Tyr11. To test the functional significance of this effect, we used NMR saturation transfer to investigate the interaction between brazzein and the amino terminal domain of the sweet receptor subunit T1R2; the results showed a stronger interaction at 7°C than at 37°C. Thus the low temperature conformation, which alters the orientations of two loops known to be critical for the sweetness of brazzein, may represent the bound state of brazzein in the complex with the human sweet receptor.
Organizational Affiliation:
National Magnetic Resonance Facility at Madison, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA.