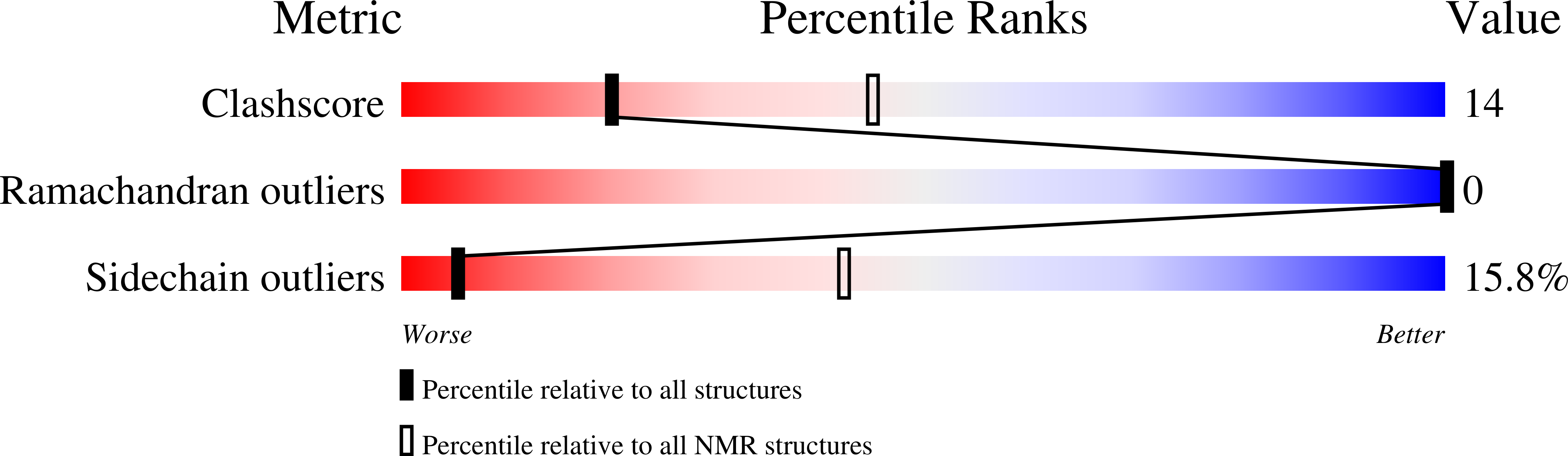Dermatophytic defensin with antiinfective potential
Zhu, S., Gao, B., Harvey, P.J., Craik, D.J.(2012) Proc Natl Acad Sci U S A 109: 8495-8500
- PubMed: 22586077
- DOI: https://doi.org/10.1073/pnas.1201263109
- Primary Citation of Related Structures:
2LR5 - PubMed Abstract:
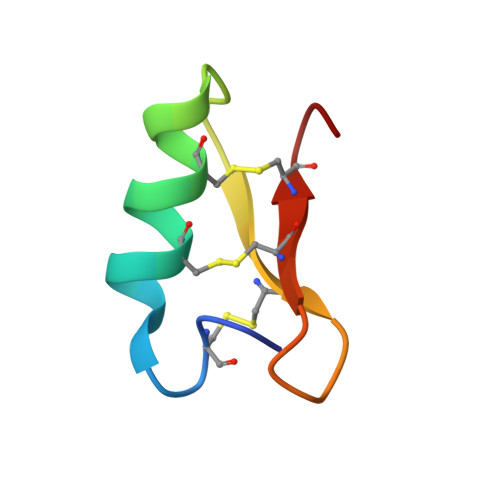
Fungi are a newly emerging source of peptide antibiotics with therapeutic potential. Here, we report 17 new fungal defensin-like peptide (fDLP) genes and the detailed characterization of a corresponding synthetic fDLP (micasin) from a dermatophyte in terms of its structure, activity and therapeutic potential. NMR analysis showed that synthetic micasin adopts a "hallmark" cysteine-stabilized α-helical and β-sheet fold. It was active on both gram-positive and gram-negative bacteria, and importantly it killed two clinical isolates of methicillin-resistant Staphylococcus aureus and the opportunistic pathogen Pseudomonas aeruginosa at low micromolar concentrations. Micasin killed approximately 100% of treated bacteria within 3 h through a membrane nondisruptive mechanism of action, and showed extremely low hemolysis and high serum stability. Consistent with these functional properties, micasin increases survival in mice infected by the pathogenic bacteria in a peritonitis model. Our work represents a valuable approach to explore novel peptide antibiotics from a large resource of fungal genomes.
Organizational Affiliation:
Group of Animal Innate Immunity, State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, 100101 Beijing, China. Zhusy@ioz.ac.cn