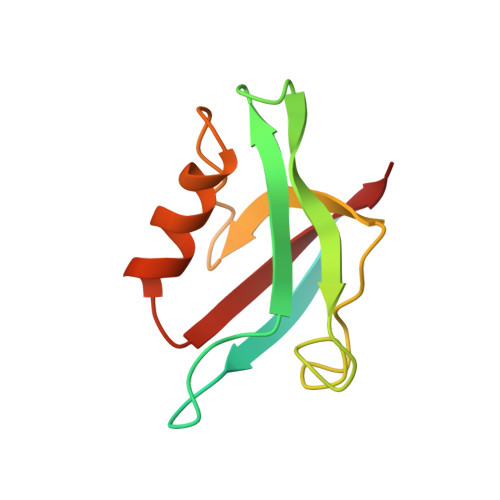
Association of the cystic fibrosis transmembrane regulator with CAL: structural features and molecular dynamics.
Piserchio, A., Fellows, A., Madden, D.R., Mierke, D.F.(2005) Biochemistry 44: 16158
- PubMed: 16331976
- DOI: https://doi.org/10.1021/bi0516475
- Primary Citation of Related Structures:
2LOB - PubMed Abstract:
The association of the cystic fibrosis transmembrane regulator (CFTR) with two PDZ-containing molecular scaffolds (CAL and EBP50) plays an important role in CFTR trafficking and membrane maintenance. The CFTR-molecular scaffold interaction is mediated by the association of the C-terminus of the transmembrane regulator with the PDZ domains. Here, we characterize the structure and dynamics of the PDZ of CAL and the complex formed with CFTR employing high-resolution NMR. On the basis of NMR relaxation data, the alpha2 helix as well as the beta2-beta3 loop of CAL PDZ domain undergoes rapid dynamics. Molecular dynamics simulations suggest a concerted motion between the alpha2 helix and the beta1-beta2 and beta2-beta3 loops, elements which define the binding pocket, suggesting that dynamics may play a role in PDZ-ligand specificity. The C-terminus of CFTR binds to CAL with the final four residues (-D(-)(3)-T-R-L(0)) within the canonical PDZ-binding motif, between the beta2 strand and the alpha2 helix. The R(-)(1) and D(-)(3) side chains make a number of contacts with the PDZ domain; many of these interactions differ from those in the CFTR-EBP50 complex, suggesting sites that can be targeted in the development of PDZ-selective inhibitors that may help modulate CFTR function.
Organizational Affiliation:
Department of Molecular Pharmacology, Division of Biology and Medicine, Brown University, Providence, Rhode Island 02912, USA.