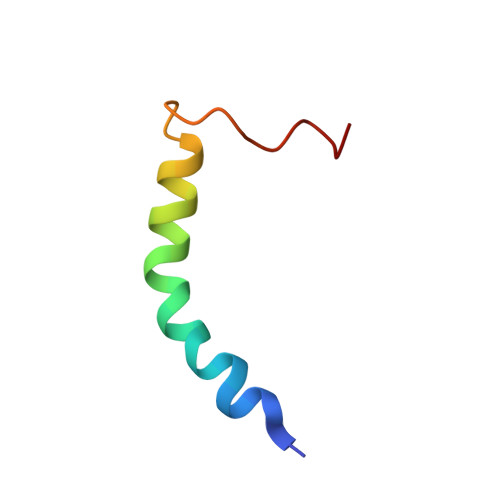
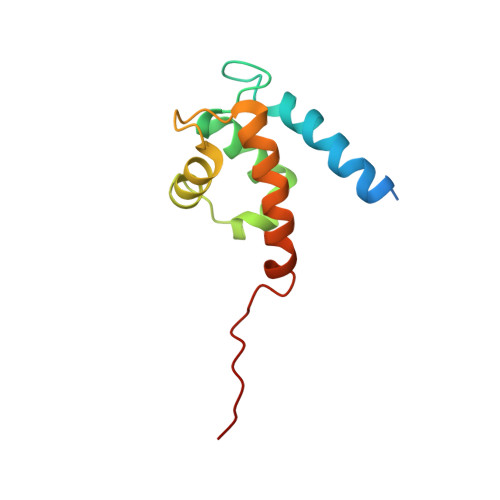
Asymmetric Mode of Ca(2+)-S100A4 Interaction with Nonmuscle Myosin IIA Generates Nanomolar Affinity Required for Filament Remodeling.
Elliott, P.R., Irvine, A.F., Jung, H.S., Tozawa, K., Pastok, M.W., Picone, R., Badyal, S.K., Basran, J., Rudland, P.S., Barraclough, R., Lian, L.Y., Bagshaw, C.R., Kriajevska, M., Barsukov, I.L.(2012) Structure 20: 654-666
- PubMed: 22483112
- DOI: https://doi.org/10.1016/j.str.2012.02.002
- Primary Citation of Related Structures:
2LNK - PubMed Abstract:
Filament assembly of nonmuscle myosin IIA (NMIIA) is selectively regulated by the small Ca²⁺-binding protein, S100A4, which causes enhanced cell migration and metastasis in certain cancers. Our NMR structure shows that an S100A4 dimer binds to a single myosin heavy chain in an asymmetrical configuration. NMIIA in the complex forms a continuous helix that stretches across the surface of S100A4 and engages the Ca²⁺-dependent binding sites of each subunit in the dimer. Synergy between these sites leads to a very tight association (K(D) ∼1 nM) that is unique in the S100 family. Single-residue mutations that remove this synergy weaken binding and ameliorate the effects of S100A4 on NMIIA filament assembly and cell spreading in A431 human epithelial carcinoma cells. We propose a model for NMIIA filament disassembly by S100A4 in which initial binding to the unstructured NMIIA tail initiates unzipping of the coiled coil and disruption of filament packing.
Organizational Affiliation:
Institute of Integrative Biology, BioSciences Building, Crown Street, University of Liverpool, Liverpool L69 7ZB, UK.