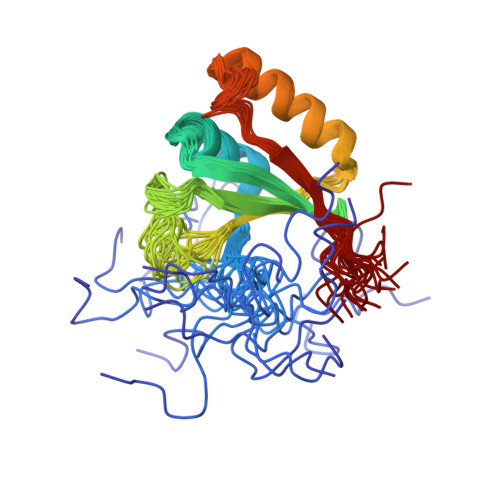
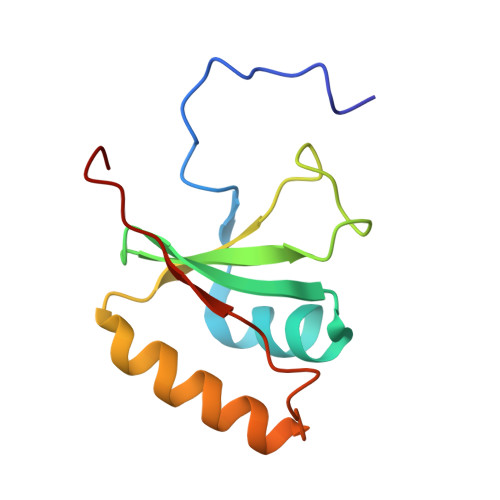
NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Ramelot, T.A., Yang, Y., Sahu, I.D., Lee, H.W., Xiao, R., Lorigan, G.A., Montelione, G.T., Kennedy, M.A.(2013) FEBS Lett 587: 3522-3528
- PubMed: 24055473
- DOI: https://doi.org/10.1016/j.febslet.2013.09.009
- Primary Citation of Related Structures:
2LNA - PubMed Abstract:
We have determined the solution NMR structure of the intermembrane space domain (IMSD) of the human mitochondrial ATPase associated with various activities (AAA) protease known as AFG3-like protein 2 (AFG3L2). Our structural analysis and molecular dynamics results indicate that the IMSD is peripherally bound to the membrane surface. This is a modification to the location of the six IMSDs in a model of the full length yeast hexaoligomeric homolog of AFG3L2 determined at low resolution by electron cryomicroscopy [1]. The predicted protein-protein interaction surface, located on the side furthest from the membrane, may mediate binding to substrates as well as prohibitins.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Northeast Structural Genomics Consortium, Miami University, Oxford, OH 45056, USA. Electronic address: theresa.ramelot@miamiOH.edu.