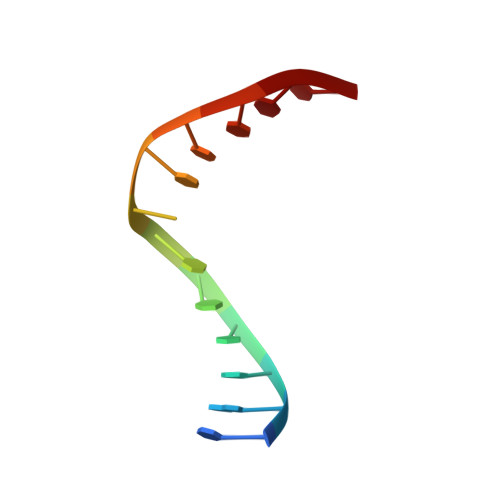
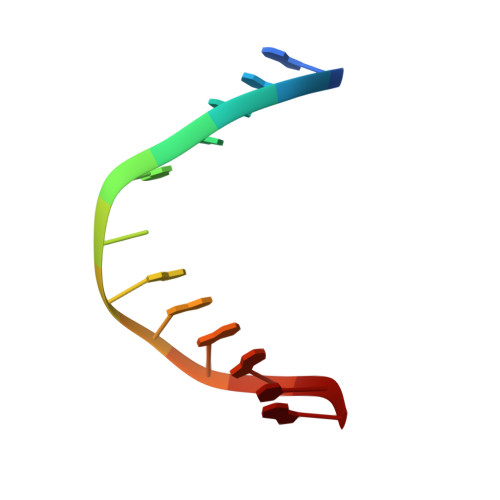
Solution structure, mechanism of replication, and optimization of an unnatural base pair.
Malyshev, D.A., Pfaff, D.A., Ippoliti, S.I., Hwang, G.T., Dwyer, T.J., Romesberg, F.E.(2010) Chemistry 16: 12650-12659
- PubMed: 20859962
- DOI: https://doi.org/10.1002/chem.201000959
- Primary Citation of Related Structures:
2LHO - PubMed Abstract:
As part of an ongoing effort to expand the genetic alphabet for in vitro and eventual in vivo applications, we have synthesized a wide variety of predominantly hydrophobic unnatural base pairs and evaluated their replication in DNA. Collectively, the results have led us to propose that these base pairs, which lack stabilizing edge-on interactions, are replicated by means of a unique intercalative mechanism. Here, we report the synthesis and characterization of three novel derivatives of the nucleotide analogue dMMO2, which forms an unnatural base pair with the nucleotide analogue d5SICS. Replacing the para-methyl substituent of dMMO2 with an annulated furan ring (yielding dFMO) has a dramatically negative effect on replication, while replacing it with a methoxy (dDMO) or with a thiomethyl group (dTMO) improves replication in both steady-state assays and during PCR amplification. Thus, dTMO-d5SICS, and especially dDMO-d5SICS, represent significant progress toward the expansion of the genetic alphabet. To elucidate the structure-activity relationships governing unnatural base pair replication, we determined the solution structure of duplex DNA containing the parental dMMO2-d5SICS pair, and also used this structure to generate models of the derivative base pairs. The results strongly support the intercalative mechanism of replication, reveal a surprisingly high level of specificity that may be achieved by optimizing packing interactions, and should prove invaluable for the further optimization of the unnatural base pair.
Organizational Affiliation:
Department of Chemistry, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.