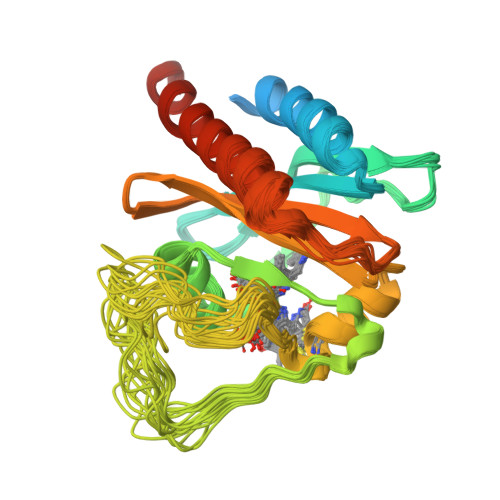
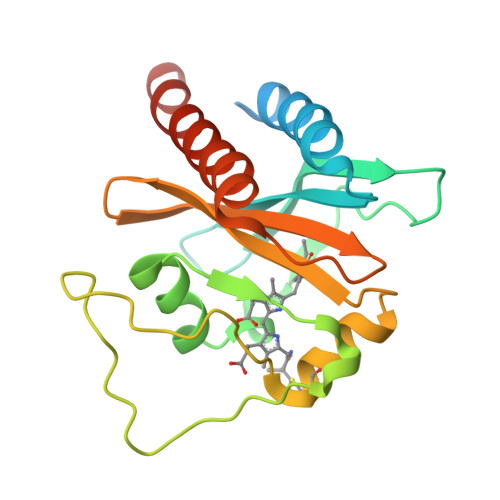
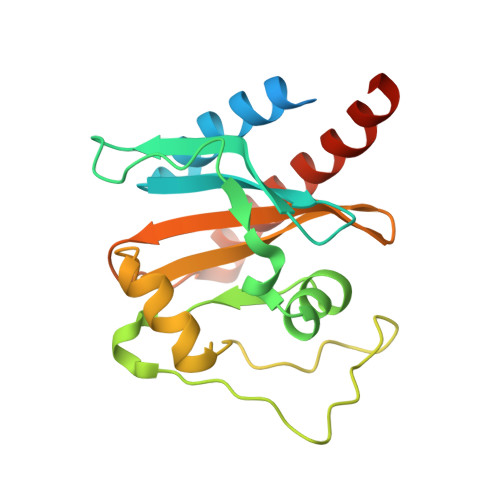
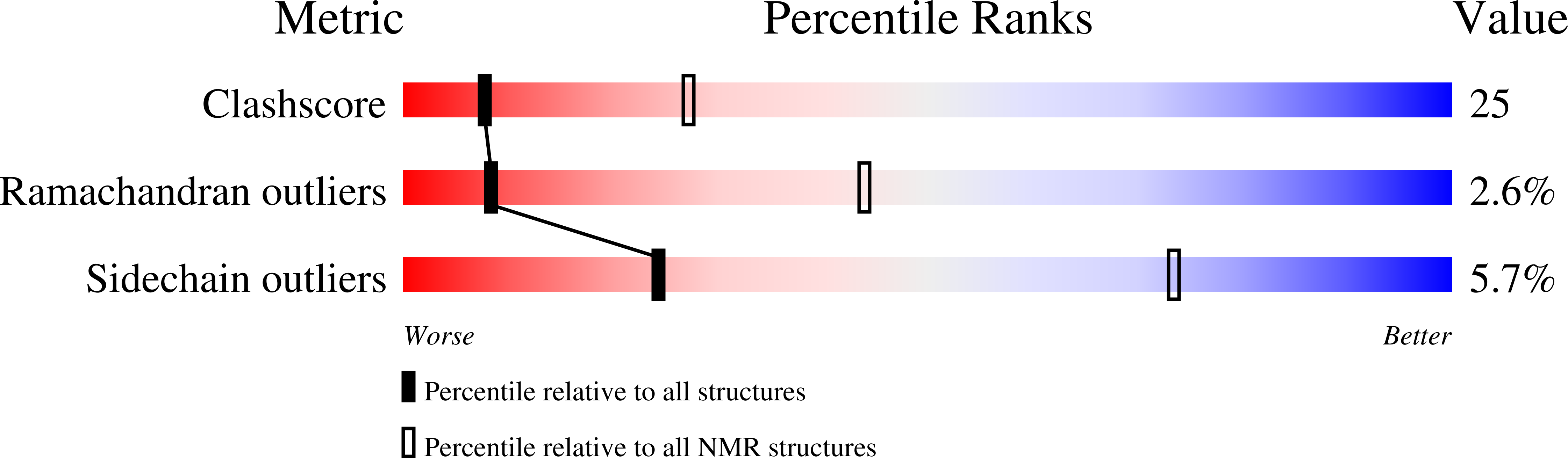Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state.
Cornilescu, G., Ulijasz, A.T., Cornilescu, C.C., Markley, J.L., Vierstra, R.D.(2008) J Mol Biology 383: 403-413
- PubMed: 18762196
- DOI: https://doi.org/10.1016/j.jmb.2008.08.034
- Primary Citation of Related Structures:
2K2N, 2LB9 - PubMed Abstract:
The unique photochromic absorption behavior of phytochromes (Phys) depends on numerous reversible interactions between the bilin chromophore and the associated polypeptide. To help define these dynamic interactions, we determined by NMR spectroscopy the first solution structure of the chromophore-binding cGMP phosphodiesterase/adenylcyclase/FhlA (GAF) domain from a cyanobacterial Phy assembled with phycocyanobilin (PCB). The three-dimensional NMR structure of Synechococcus OS-B' cyanobacterial Phy 1 in the red-light-absorbing state of Phy (Pr) revealed that PCB is bound to Cys138 of the GAF domain via the A-ring ethylidene side chain and is buried within the GAF domain in a ZZZsyn,syn,anti configuration. The D ring of the chromophore sits within a hydrophobic pocket and is tilted by approximately 80 degrees relative to the B/C rings by contacts with Lys52 and His169. The solution structure revealed remarkable flexibility for PCB and several adjacent amino acids, indicating that the Pr chromophore has more freedom in the binding pocket than anticipated. The propionic acid side chains of rings B and C and Arg101 and Arg133 nearby are especially mobile and can assume several distinct and energetically favorable conformations. Mutagenic studies on these arginines, which are conserved within the Phy superfamily, revealed that they have opposing roles, with Arg101 and Arg133 helping stabilize and destabilize the far-red-light-absorbing state of Phy (Pfr), respectively. Given the fact that the Synechococcus OS-B' GAF domain can, by itself, complete the Pr --> Pfr photocycle, it should now be possible to determine the solution structure of the Pfr chromophore and surrounding pocket using this Pr structure as a framework.
Organizational Affiliation:
National Magnetic Resonance Facility at Madison, University of Wisconsin, Madison, WI 53706, USA.