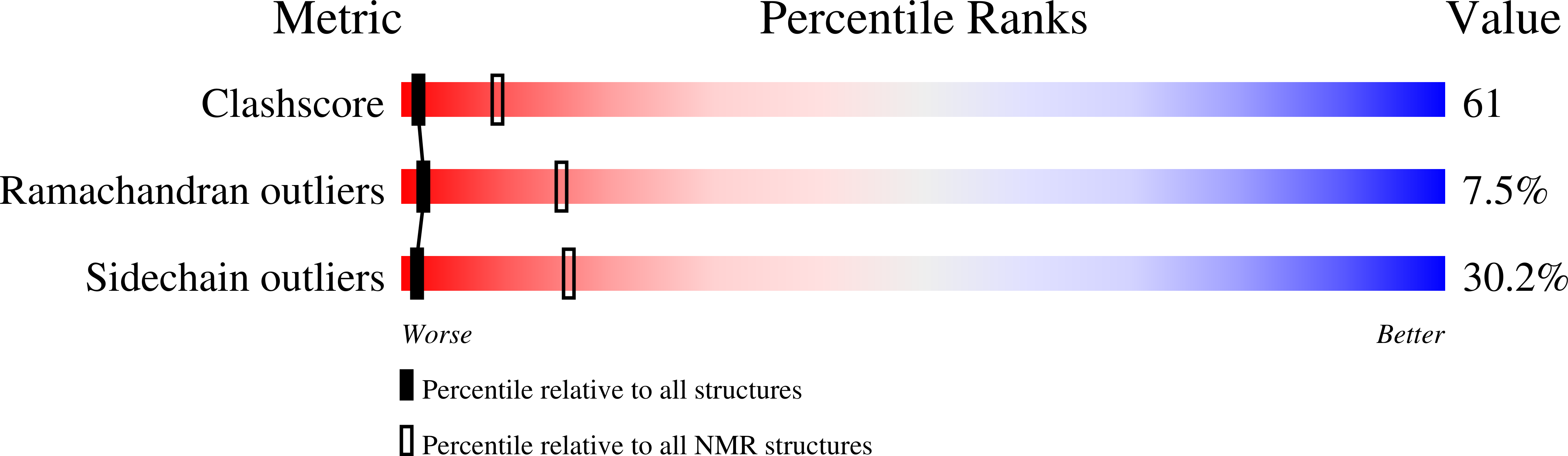Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
Watanabe, Y., Noda, N.N., Kumeta, H., Suzuki, K., Ohsumi, Y., Inagaki, F.(2010) J Biol Chem 285: 30026-30033
- PubMed: 20659891
- DOI: https://doi.org/10.1074/jbc.M110.143545
- Primary Citation of Related Structures:
2KZB, 2KZK - PubMed Abstract:
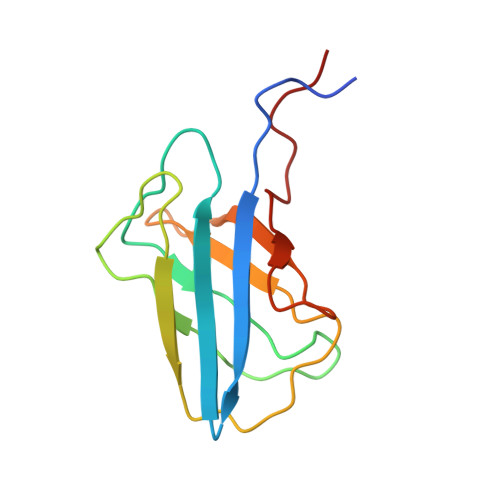
In the yeast Saccharomyces cerevisiae, a precursor form of aminopeptidase I (prApe1) and α-mannosidase (Ams1) are selectively transported to the vacuole through the cytoplasm-to-vacuole targeting pathway under vegetative conditions and through autophagy under starvation conditions. Atg19 plays a central role in these processes by linking Ams1 and prApe1 to Atg8 and Atg11. However, little is known about the molecular mechanisms of cargo recognition by Atg19. Here, we report structural and functional analyses of Atg19 and its paralog, Atg34. A protease-resistant domain was identified in the C-terminal region of Atg19, which was also conserved in Atg34. In vitro pulldown assays showed that the C-terminal domains of both Atg19 and Atg34 are responsible for Ams1 binding; these domains are hereafter referred to as Ams1-binding domains (ABDs). The transport of Ams1, but not prApe1, was blocked in atg19Δatg34Δ cells expressing Atg19(ΔABD), indicating that ABD is specifically required for Ams1 transport. We then determined the solution structures of the ABDs of Atg19 and Atg34 using NMR spectroscopy. Both ABD structures have a canonical immunoglobulin fold consisting of eight β-strands with highly conserved loops clustered at one side of the fold. These facts, together with the results of a mutational analysis, suggest that ABD recognizes Ams1 using these conserved loops.
Organizational Affiliation:
Department of Structural Biology, Graduate School of Pharmaceutical Sciences, Hokkaido University, N-12, W-6 Kita-ku, Sapporo 060-0812, Japan.