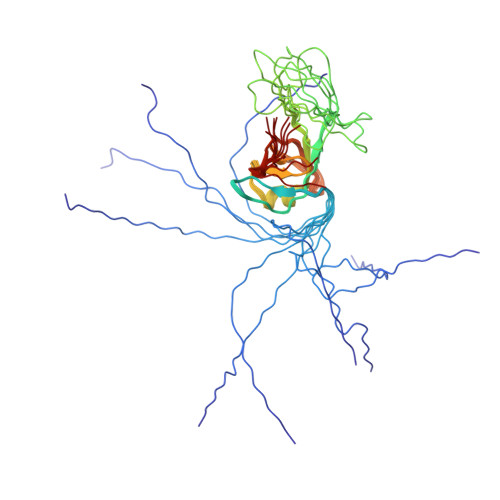
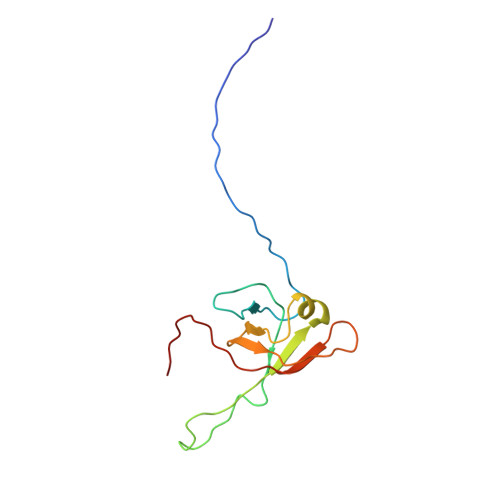
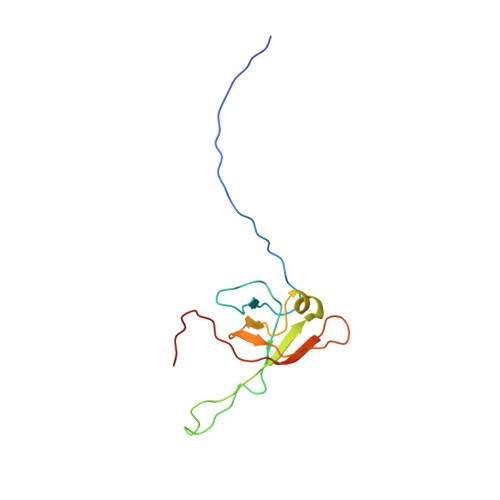
Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Cardarelli, L., Pell, L.G., Neudecker, P., Pirani, N., Liu, A., Baker, L.A., Rubinstein, J.L., Maxwell, K.L., Davidson, A.R.(2010) Proc Natl Acad Sci U S A 107: 14384-14389
- PubMed: 20660769
- DOI: https://doi.org/10.1073/pnas.1005822107
- Primary Citation of Related Structures:
2KX4 - PubMed Abstract:
Evolutionary relationships may exist among very diverse groups of proteins even though they perform different functions and display little sequence similarity. The tailed bacteriophages present a uniquely amenable system for identifying such groups because of their huge diversity yet conserved genome structures. In this work, we used structural, functional, and genomic context comparisons to conclude that the head-tail connector protein and tail tube protein of bacteriophage lambda diverged from a common ancestral protein. Further comparisons of tertiary and quaternary structures indicate that the baseplate hub and tail terminator proteins of bacteriophage may also be part of this same family. We propose that all of these proteins evolved from a single ancestral tail tube protein fold, and that gene duplication followed by differentiation led to the specialized roles of these proteins seen in bacteriophages today. Although this type of evolutionary mechanism has been proposed for other systems, our work provides an evolutionary mechanism for a group of proteins with different functions that bear no sequence similarity. Our data also indicate that the addition of a structural element at the N terminus of the lambda head-tail connector protein endows it with a distinctive protein interaction capability compared with many of its putative homologues.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, Toronto, ON, Canada M5S 1A8.