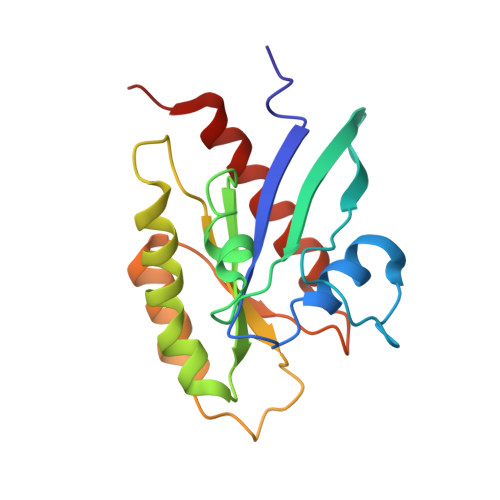
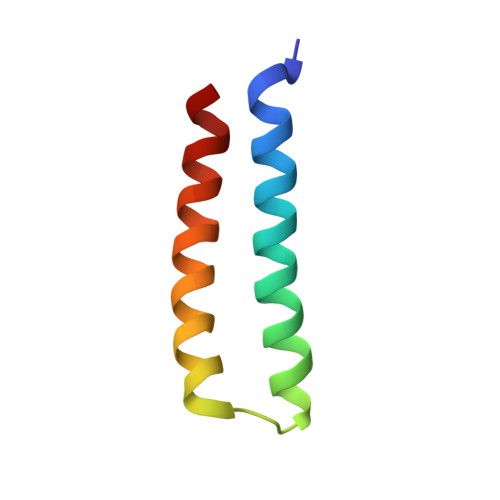
The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Fenwick, R.B., Campbell, L.J., Rajasekar, K., Prasannan, S., Nietlispach, D., Camonis, J., Owen, D., Mott, H.R.(2010) Structure 18: 985-995
- PubMed: 20696399
- DOI: https://doi.org/10.1016/j.str.2010.05.013
- Primary Citation of Related Structures:
2KWH, 2KWI - PubMed Abstract:
RLIP76 (RalBP1) is a multidomain protein that interacts with multiple small G protein families: Ral via a specific binding domain, and Rho and R-Ras via a GTPase activating domain. RLIP76 interacts with endocytosis proteins and has also been shown to behave as a membrane ATPase that transports chemotherapeutic agents from the cell. We have determined the structure of the Ral-binding domain of RLIP76 and show that it comprises a coiled-coil motif. The structure of the RLIP76-RalB complex reveals a novel mode of binding compared to the structures of RalA complexed with the exocyst components Sec5 and Exo84. RLIP76 interacts with both nucleotide-sensitive regions of RalB, and key residues in the interface have been identified using affinity measurements of RalB mutants. Sec5, Exo84, and RLIP76 bind Ral proteins competitively and with similar affinities in vitro.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, UK.