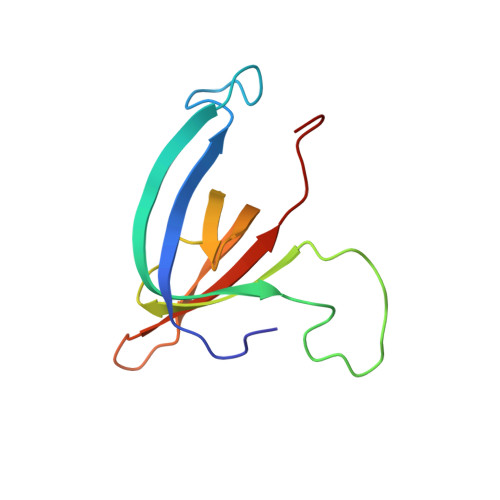
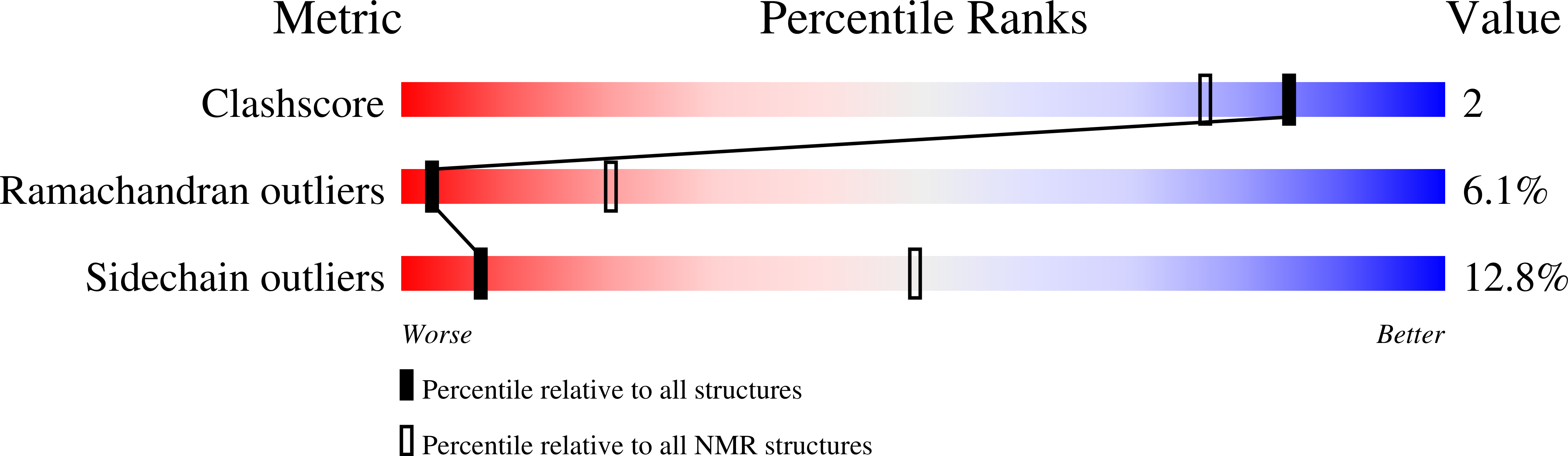Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating.
Lhuillier, S., Gallopin, M., Gilquin, B., Brasiles, S., Lancelot, N., Letellier, G., Gilles, M., Dethan, G., Orlova, E.V., Couprie, J., Tavares, P., Zinn-Justin, S.(2009) Proc Natl Acad Sci U S A 106: 8507-8512
- PubMed: 19433794
- DOI: https://doi.org/10.1073/pnas.0812407106
- Primary Citation of Related Structures:
2KBZ, 2KCA - PubMed Abstract:
In many bacterial viruses and in certain animal viruses, the double-stranded DNA genome enters and exits the capsid through a portal gatekeeper. We report a pseudoatomic structure of a complete portal system. The bacteriophage SPP1 gatekeeper is composed of dodecamers of the portal protein gp6, the adaptor gp15, and the stopper gp16. The solution structures of gp15 and gp16 were determined by NMR. They were then docked together with the X-ray structure of gp6 into the electron density of the approximately 1-MDa SPP1 portal complex purified from DNA-filled capsids. The resulting structure reveals that gatekeeper assembly is accompanied by a large rearrangement of the gp15 structure and by folding of a flexible loop of gp16 to form an intersubunit parallel beta-sheet that closes the portal channel. This stopper system prevents release of packaged DNA. Disulfide cross-linking between beta-strands of the stopper blocks the key conformational changes that control genome ejection from the virus at the beginning of host infection.
Organizational Affiliation:
Unité de Virologie Moléculaire et Structurale, Centre National de la Recherche Scientifique, Unité Mixte de Recherche 2472, Institut National de la Recherche Agronomique, 91198 Gif-sur-Yvette, France.