The Structure of Binder of Arl2 (BART) Reveals a Novel G Protein Binding Domain: IMPLICATIONS FOR FUNCTION.
Bailey, L.K., Campbell, L.J., Evetts, K.A., Littlefield, K., Rajendra, E., Nietlispach, D., Owen, D., Mott, H.R.(2009) J Biol Chem 284: 992-999
- PubMed: 18981177
- DOI: https://doi.org/10.1074/jbc.M806167200
- Primary Citation of Related Structures:
2K9A - PubMed Abstract:
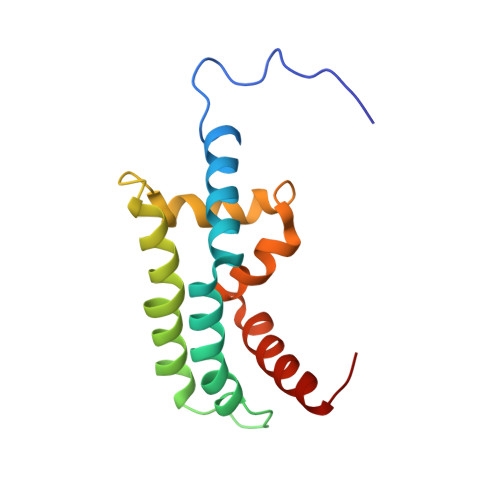
The ADP-ribosylation factor-like (Arl) family of small G proteins are involved in the regulation of diverse cellular processes. Arl2 does not appear to be membrane localized and has been implicated as a regulator of microtubule dynamics. The downstream effector for Arl2, Binder of Arl 2 (BART) has no known function but, together with Arl2, can enter mitochondria and bind the adenine nucleotide transporter. We have solved the solution structure of BART and show that it forms a novel fold composed of six alpha-helices that form three interlocking "L" shapes. Analysis of the backbone dynamics reveals that the protein is highly anisotropic and that the loops between the central helices are dynamic. The regions involved in the binding of Arl2 were mapped onto the surface of BART and are found to localize to these loop regions. BART has faces of differing charge and structural elements, which may explain how it can interact with other proteins.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, United Kingdom.