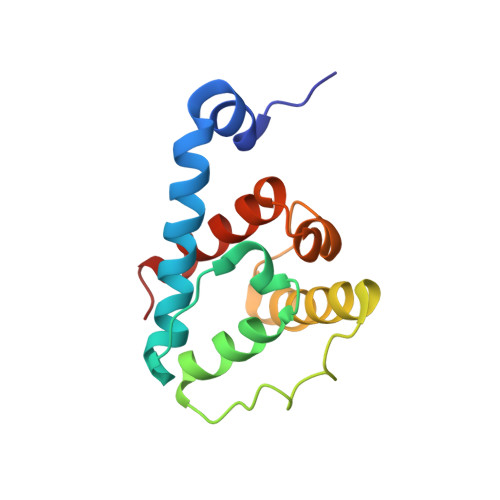
The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
Wang, X., Fukuda, K., Byeon, I.J., Velyvis, A., Wu, C., Gronenborn, A., Qin, J.(2008) J Biol Chem 283: 21113-21119
- PubMed: 18508764
- DOI: https://doi.org/10.1074/jbc.M801270200
- Primary Citation of Related Structures:
2K2R - PubMed Abstract:
Alpha-parvin is an essential component of focal adhesions (FAs), which are large multiprotein complexes that link the plasma membrane and actin cytoskeleton. Alpha-parvin contains two calponin homology (CH) domains and its C-terminal CH2 domain binds multiple targets including paxillin LD motifs for regulating the FA network and signaling. Here we describe the solution structure of alpha-parvin CH2 bound to paxillin LD1. We show that although CH2 contains the canonical CH-fold, a previously defined N-terminal linker forms an alpha-helix that packs unexpectedly with the C-terminal helix of CH2, resulting in a novel variant of the CH domain. Importantly, such packing generates a hydrophobic surface that recognizes the Leu-rich face of paxillin-LD1, and the binding pattern differs drastically from the classical paxillin-LD binding to four-helix bundle proteins such as focal adhesion kinase. These results define a novel modular recognition mode and reveal how alpha-parvin associates with paxillin to mediate the FA assembly and signaling.
Organizational Affiliation:
Structural Biology Program, Lerner Research Institute, Cleveland Clinic, 9500 Euclid Avenue, Cleveland, OH 44195, USA.