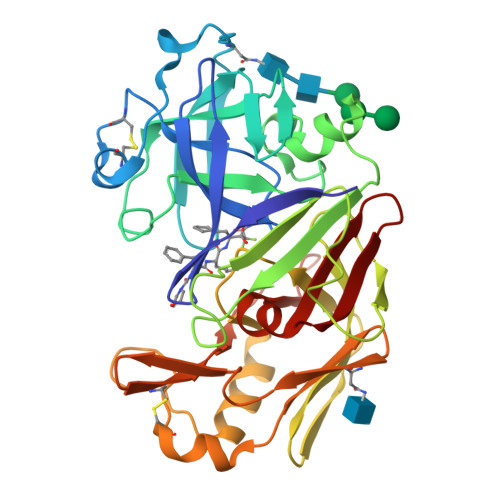
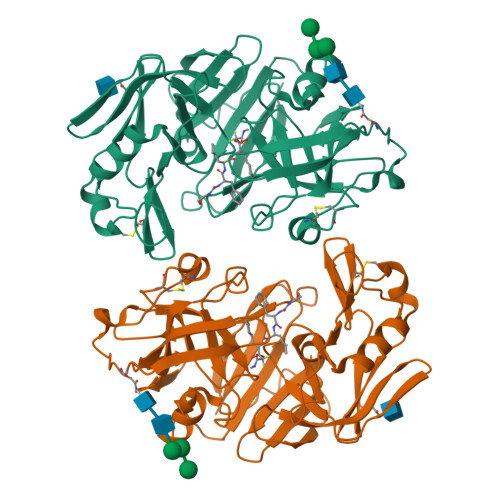
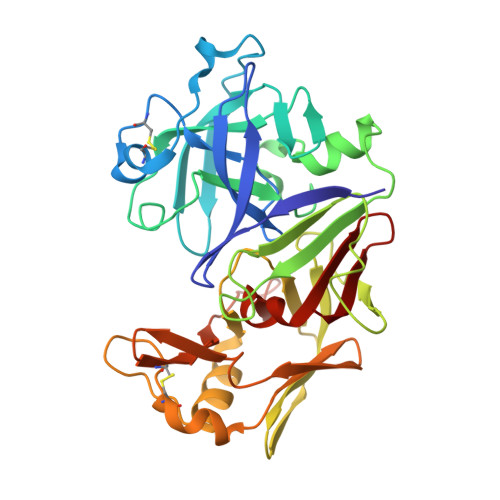
The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
Aguilar, C.F., Cronin, N.B., Badasso, M., Dreyer, T., Newman, M.P., Cooper, J.B., Hoover, D.J., Wood, S.P., Johnson, M.S., Blundell, T.L.(1997) J Mol Biology 267: 899-915
- PubMed: 9135120
- DOI: https://doi.org/10.1006/jmbi.1996.0880
- Primary Citation of Related Structures:
2JXR - PubMed Abstract:
The crystal structures of glycosylated native proteinase A, an aspartic proteinase found in the vacuole of Saccharomyces cerevisiae, and its complex with a difluorostatone-containing tripeptide have been determined by molecular replacement to 3.5 A and 2.4 A resolutions, respectively. Superposition of the bound and native forms gave an r.m.s. difference of 0.6 A largely reflecting the poor resolution of the native crystal structure. The secondary and tertiary structures are highly similar to those found in porcine pepsin and lysosomal cathepsin D; superposition of the structure of proteinase A bound to the difluorostatone inhibitor on those of pepsin and cathepsin D gave pairwise r.m.s. differences for C(alpha) atoms of 1.36 A and 0.88 A. Most differences occur in loop regions. Comparison of the structure of the proteinase A-difluorostatone complex with that of endothiapepsin bound with the same inhibitor shows that the conformation and hydrogen bond interactions of the inhibitor in the active site are very similar, even though the enzymes have only 27% sequence identity. Electron density for the crystal structure of the proteinase A complex reveals five residues of the oligosaccharide structure attached to Asn67: Man-(1 --> 2)-alpha-Man-(1 --> 3)-beta-Man-(1 --> 4)-beta-GlcNAc-(1 --> 4)-beta-GlcNAc-Asn-67. The first three residues of the oligosaccharide cover the same region of the protein surface as those of the oligosaccharide attached to the equivalent position in cathepsin D. The second carbohydrate attachment site is disordered beyond the first carbohydrate residue in both enzymes.
Organizational Affiliation:
Department of Crystallography, Birkbeck College, London, UK.