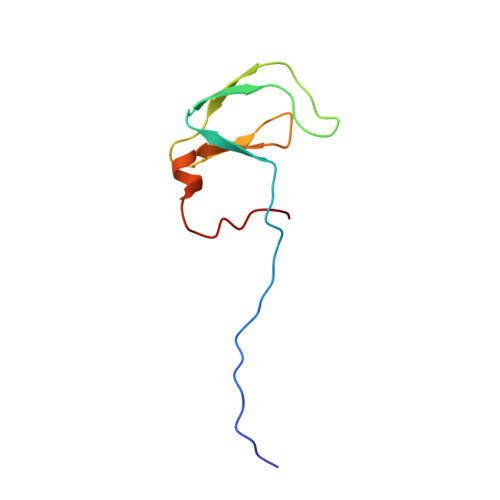
Solution NMR Structure of Rhodobacter sphaeroides protein RHOS4_26430.
Eletsky, A., Sukumaran, D., Zhang, Q., Parish, D., Xu, D., Wang, H., Janjua, H., Owens, L., Xiao, R., Liu, J., Baran, M.C., Swapna, G.V.T., Acton, T.B., Rost, B., Montelione, G.T., Szyperski, T.To be published.