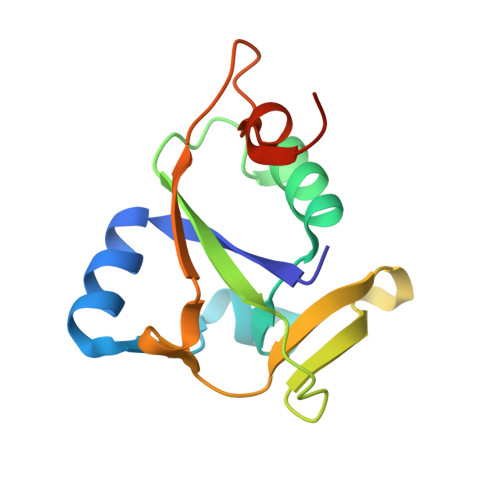
Solution NMR structure of CC0527 from Caulobacter crescentus
Aramini, J.M., Rossi, P., Moseley, H.N.B., Wang, D., Nwosu, C., Cunningham, K., Ma, L., Xiao, R., Liu, J., Baran, M.C., Swapna, G.V.T., Acton, T.B., Rost, B., Montelione, G.T.To be published.