1,3-Disubstituted 4-Aminopiperidines as Useful Tools in the Optimization of the 2-Aminobenzo[A]Quinolizine Dipeptidyl Peptidase Iv Inhibitors.
Lubbers, T., Bohringer, M., Gobbi, L., Hennig, M., Hunziker, D., Kuhn, B., Loffler, B., Mattei, P., Narquizian, R., Peters, J., Ruff, Y., Wessel, H.P., Wyss, P.(2007) Bioorg Med Chem Lett 17: 2966
- PubMed: 17418568
- DOI: https://doi.org/10.1016/j.bmcl.2007.03.072
- Primary Citation of Related Structures:
2JID - PubMed Abstract:
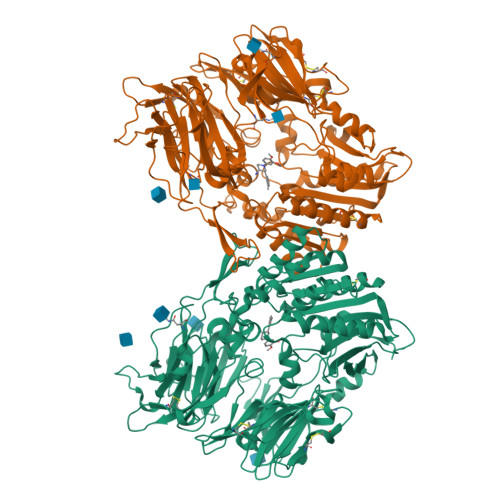
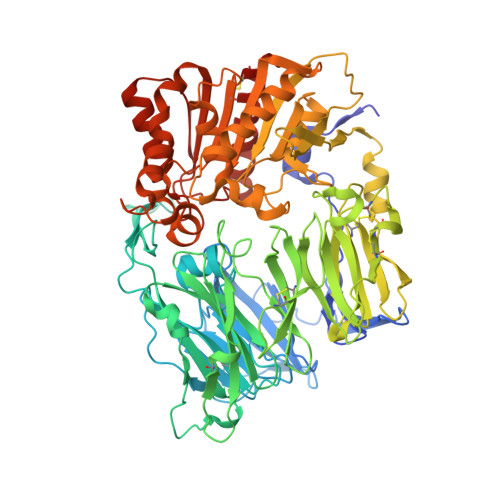
In a search for novel DPP-IV inhibitors, 2-aminobenzo[a]quinolizines were identified as submicromolar HTS hits. Due to the difficult synthetic access to this compound class, 1,3-disubstituted 4-aminopiperidines were used as model compounds for optimization. The developed synthetic methodology and the SAR could be transferred to the 2-aminobenzo[a]quinolizine series, leading to highly active DPP-IV inhibitors.
Organizational Affiliation:
Discovery Research, Pharmaceutical Division, F. Hoffmann-La Roche Ltd, CH-4070 Basel, Switzerland. thomas.luebbers@roche.com