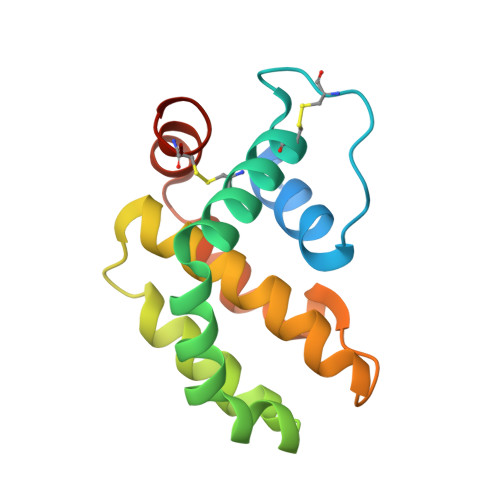
A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak.
Kvansakul, M., van Delft, M.F., Lee, E.F., Gulbis, J.M., Fairlie, W.D., Huang, D.C., Colman, P.M.(2007) Mol Cell 25: 933-942
- PubMed: 17386268
- DOI: https://doi.org/10.1016/j.molcel.2007.02.004
- Primary Citation of Related Structures:
2JBX, 2JBY - PubMed Abstract:
Many viruses express antiapoptotic proteins to counter host defense mechanisms that would otherwise trigger the rapid clearance of infected cells. For example, adenoviruses and some gamma-herpesviruses express homologs of prosurvival Bcl-2 to subvert the host's apoptotic machinery. Myxoma virus, a double-stranded DNA virus of the pox family, harbors antiapoptotic M11L, its virulence factor. Analysis of its three-dimensional structure reveals that despite lacking any primary sequence similarity to Bcl-2, it adopts a virtually identical protein fold. This allows it to associate with BH3 domains, especially those of Bax and Bak. We found that M11L acts primarily by sequestering Bax and Bak, thereby blocking the killing action of these essential cell-death mediators. These findings expand the family of protein sequences that act like Bcl-2 to block apoptosis and support the conclusion that the prosurvival action of these proteins critically depends on their ability to bind and antagonize Bax and/or Bak.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3050, Australia.