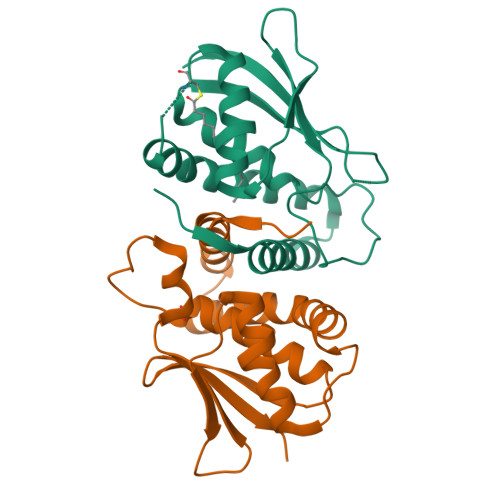
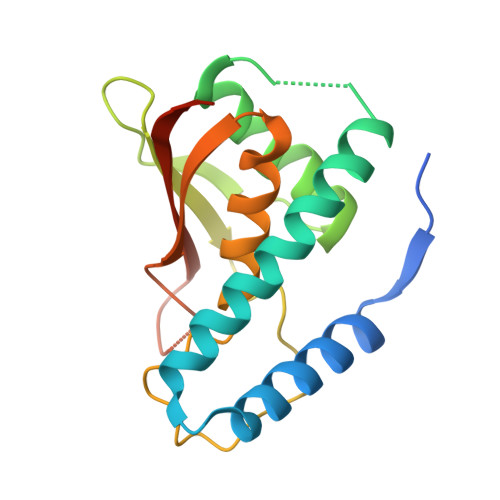
The Architecture of the Multisubunit Trapp I Complex Suggests a Model for Vesicle Tethering.
Kim, Y.-G., Raunser, S., Munger, C., Wagner, J., Song, Y.-L., Cygler, M., Walz, T., Oh, B.-H., Sacher, M.(2006) Cell 127: 817
- PubMed: 17110339
- DOI: https://doi.org/10.1016/j.cell.2006.09.029
- Primary Citation of Related Structures:
2J3R, 2J3T, 2J3W - PubMed Abstract:
Transport protein particle (TRAPP) I is a multisubunit vesicle tethering factor composed of seven subunits involved in ER-to-Golgi trafficking. The functional mechanism of the complex and how the subunits interact to form a functional unit are unknown. Here, we have used a multidisciplinary approach that includes X-ray crystallography, electron microscopy, biochemistry, and yeast genetics to elucidate the architecture of TRAPP I. The complex is organized through lateral juxtaposition of the subunits into a flat and elongated particle. We have also localized the site of guanine nucleotide exchange activity to a highly conserved surface encompassing several subunits. We propose that TRAPP I attaches to Golgi membranes with its large flat surface containing many highly conserved residues and forms a platform for protein-protein interactions. This study provides the most comprehensive view of a multisubunit vesicle tethering complex to date, based on which a model for the function of this complex, involving Rab1-GTP and long, coiled-coil tethers, is presented.
Organizational Affiliation:
Center for Biomolecular Recognition and Division of Molecular and Life Sciences, Department of Life Sciences, Pohang University of Science and Technology, Pohang, Kyungbuk 790-784, South Korea.