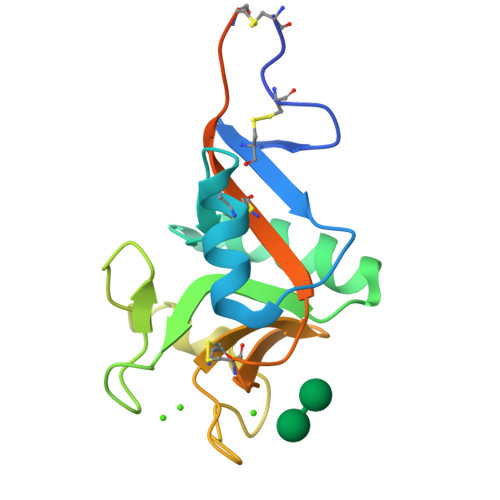
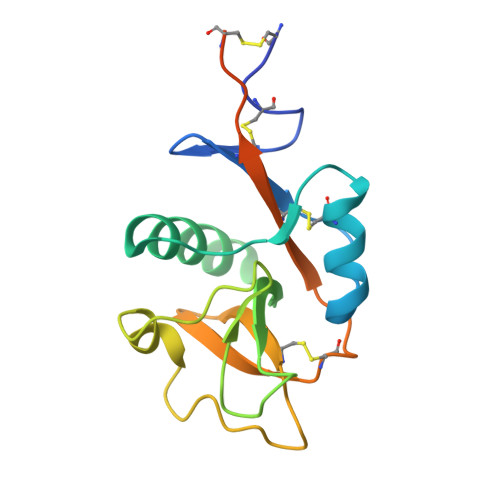
Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins.
Feinberg, H., Castelli, R., Drickamer, K., Seeberger, P.H., Weis, W.I.(2007) J Biological Chem 282: 4202-4209
- PubMed: 17150970
- DOI: https://doi.org/10.1074/jbc.M609689200
- Primary Citation of Related Structures:
2IT5, 2IT6 - PubMed Abstract:
The dendritic cell surface receptor DC-SIGN and the closely related endothelial cell receptor DC-SIGNR specifically recognize high mannose N-linked carbohydrates on viral pathogens. Previous studies have shown that these receptors bind the outer trimannose branch Manalpha1-3[Manalpha1-6]Manalpha present in high mannose structures. Although the trimannoside binds to DC-SIGN or DC-SIGNR more strongly than mannose, additional affinity enhancements are observed in the presence of one or more Manalpha1-2Manalpha moieties on the nonreducing termini of oligomannose structures. The molecular basis of this enhancement has been investigated by determining crystal structures of DC-SIGN bound to a synthetic six-mannose fragment of a high mannose N-linked oligosaccharide, Manalpha1-2Manalpha1-3[Manalpha1-2Manalpha1-6]Manalpha1-6Man and to the disaccharide Manalpha1-2Man. The structures reveal mixtures of two binding modes in each case. Each mode features typical C-type lectin binding at the principal Ca2+-binding site by one mannose residue. In addition, other sugar residues form contacts unique to each binding mode. These results suggest that the affinity enhancement displayed toward oligosaccharides decorated with the Manalpha1-2Manalpha structure is due in part to multiple binding modes at the primary Ca2+ site, which provide both additional contacts and a statistical (entropic) enhancement of binding.
Organizational Affiliation:
Departments of Structural Biology and Molecular & Cellular Physiology, Stanford University School of Medicine, Stanford, California 94305, USA.