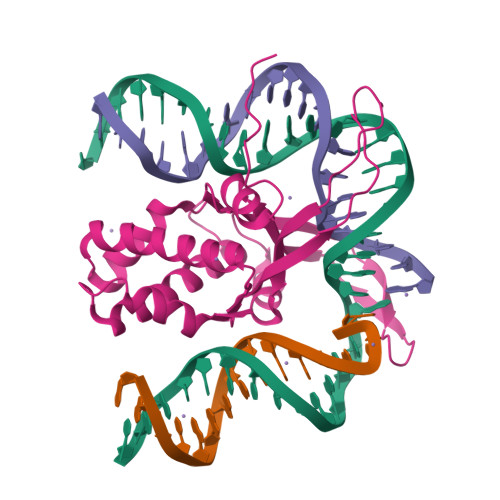
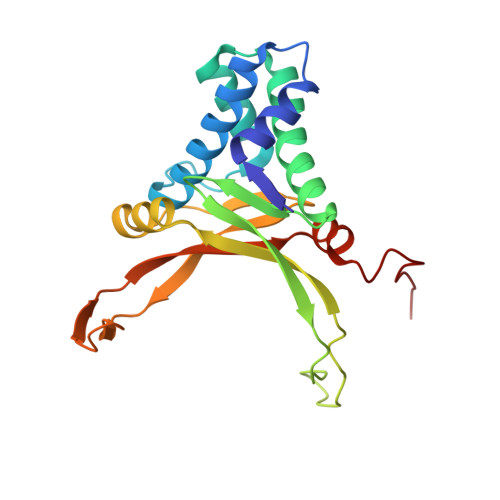
A Divalent Metal-mediated Switch Controlling Protein-induced DNA Bending
Bao, Q., Chen, H., Liu, Y., Yan, J., Droge, P., Davey, C.A.(2007) J Mol Biology 367: 731-740
- PubMed: 17276457
- DOI: https://doi.org/10.1016/j.jmb.2006.09.082
- Primary Citation of Related Structures:
2IIE, 2IIF - PubMed Abstract:
Architectural proteins that reconfigure the paths of DNA segments are required for the establishment of functional interfaces in many genomic transactions. A single-chain derivative of the DNA architectural protein integration host factor was found to adopt two stable conformational states in complex with a specific DNA target. In the so-called open state, the degree of protein-induced DNA bending is reduced significantly compared with the closed state. The conformational switch between these states is controlled by divalent metal binding in two electronegative zones arising from the lysine-to-glutamate substitution in the protein body proximal to the phosphate backbone of one DNA arm. We show that this switch can be employed to control the efficiency of site-specific recombination catalyzed by lambda integrase. Introduction of acidic residues at the protein-DNA interface holds potential for the design of metal-mediated switches for the investigation of functional relationships.
Organizational Affiliation:
Division of Genomics and Genetics, School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore.