Influence of the target base partner on the methylation rate of the adenine-specific DNA methyltransferase M.TaqI
Lenz, T., Scheidig, A.J., Weinhold, E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Modification methylase TaqI | E [auth A], F [auth D] | 421 | Thermus aquaticus | Mutation(s): 0 Gene Names: taqIM EC: 2.1.1.72 | 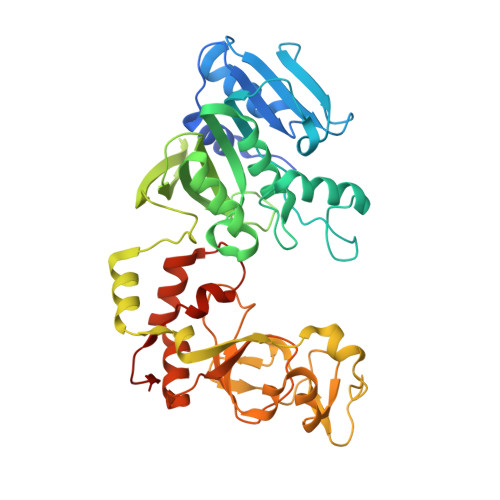 |
UniProt | |||||
Find proteins for P14385 (Thermus aquaticus) Explore P14385 Go to UniProtKB: P14385 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14385 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*GP*TP*TP*CP*GP*AP*TP*GP*TP*C)-3' | A [auth B], C [auth E] | 10 | N/A |  | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*GP*AP*CP*AP*(5PY)P*CP*GP*(6MA)P*AP*C)-3' | B [auth C], D [auth F] | 10 | N/A | 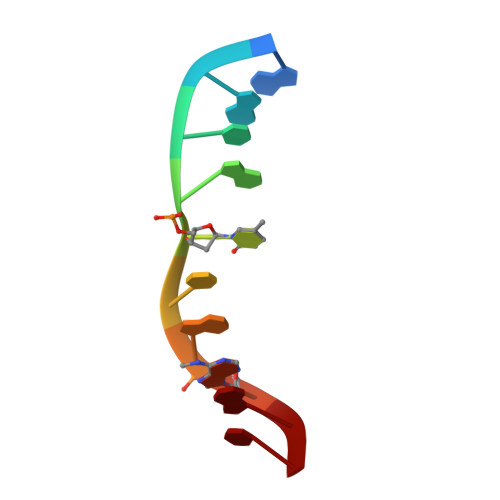 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NEA Query on NEA | H [auth A], J [auth D] | 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE C12 H18 N6 O3 S APAPOJUCRZTCHD-WOUKDFQISA-N |  | ||
| GOL Query on GOL | G [auth C], I [auth A], K [auth D], L [auth D], M [auth D] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.439 | α = 90 |
| b = 68.875 | β = 92.27 |
| c = 114.349 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| ProDC | data collection |
| MAR345 | data collection |
| XDS | data scaling |
| MOLREP | phasing |