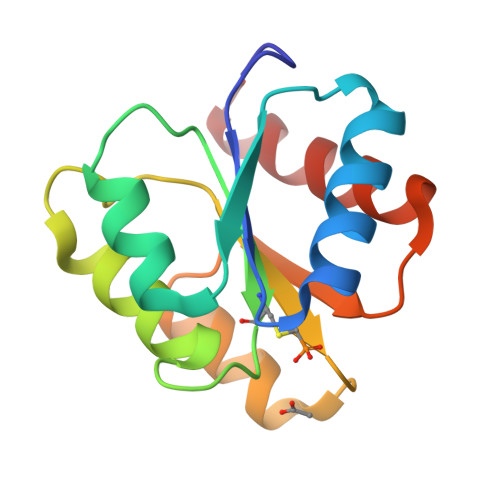
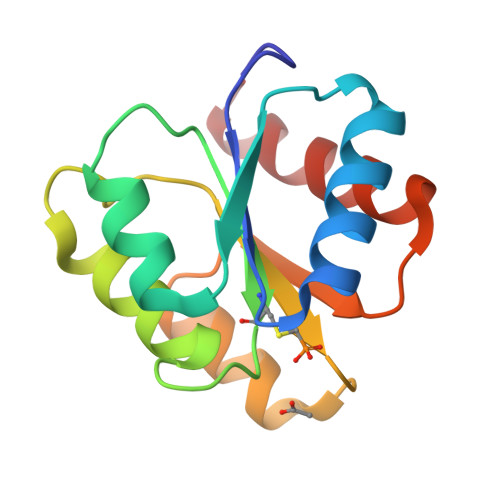
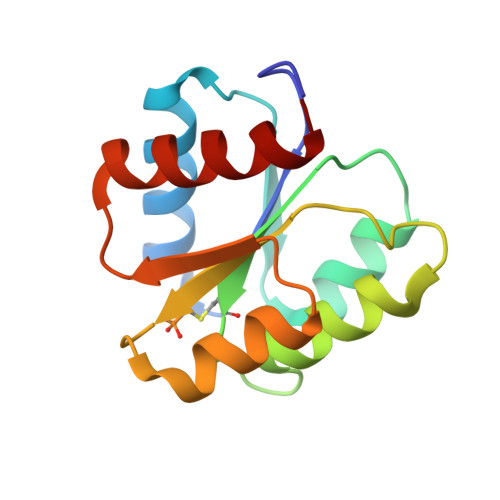
The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
McAdams, K., Casper, E.S., Matthew Haas, R., Santarsiero, B.D., Eggler, A.L., Mesecar, A., Halkides, C.J.(2008) Arch Biochem Biophys 479: 105-113
- PubMed: 18801331
- DOI: https://doi.org/10.1016/j.abb.2008.08.019
- Primary Citation of Related Structures:
2ID7, 2ID9, 2IDM - PubMed Abstract:
CheY is a response regulator in bacterial chemotaxis. Escherichia coli CheY mutants T87I and T87I/Y106W CheY are phosphorylatable on Asp57 but unable to generate clockwise rotation of the flagella. To understand this phenotype in terms of structure, stable analogs of the two CheY-P mutants were synthesized: T87I phosphono-CheY and T87I phosphono-CheY. Dissociation constants for peptides derived from flagellar motor protein FliM and phosphatase CheZ were determined for phosphono-CheY and the two mutants. The peptides bind phosphono-CheY almost as strongly as CheY-P; however, they do not bind T87I phosphono-CheY or T87I/Y106W phosphono-CheY, implying that the mutant proteins cannot bind FliM or CheZ tightly in vivo. The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY were solved to resolutions of 1.8 and 2.4A, respectively. The increased bulk of I87 forces the side-chain of Y106 or W106, into a more solvent-accessible conformation, which occludes the peptide-binding site.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of North Carolina Wilmington, 601 S. College Road, Wilmington, NC 28403, USA.