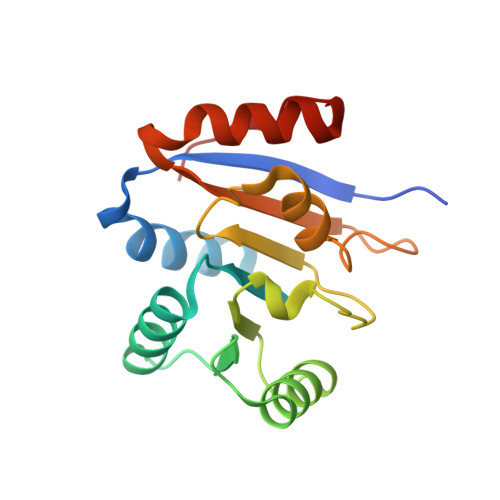
Structure of the second domain of the Bacillus subtilis DEAD-box RNA helicase YxiN.
Caruthers, J.M., Hu, Y., McKay, D.B.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 1191-1195
- PubMed: 17142894
- DOI: https://doi.org/10.1107/S1744309106044642
- Primary Citation of Related Structures:
2HJV - PubMed Abstract:
The Bacillus subtilis RNA helicase YxiN is a modular three-domain protein. The first two domains form a conserved helicase core that couples an ATPase activity to an RNA duplex-destabilization activity, while the third domain recognizes a stem-loop of 23S ribosomal RNA with high affinity and specificity. The structure of the second domain, amino-acid residues 207-368, has been solved to 1.95 A resolution, revealing a parallel alphabeta-fold. The crystallographic asymmetric unit contains two protomers; superposition shows that they differ substantially in two segments of peptide that overlap the conserved helicase sequence motifs V and VI, while the remainder of the domain is isostructural. The conformational variability of these segments suggests that induced fit is intrinsic to the recognition of ligands (ATP and RNA) and the coupling of the ATPase activity to conformational changes.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, California 94305, USA.