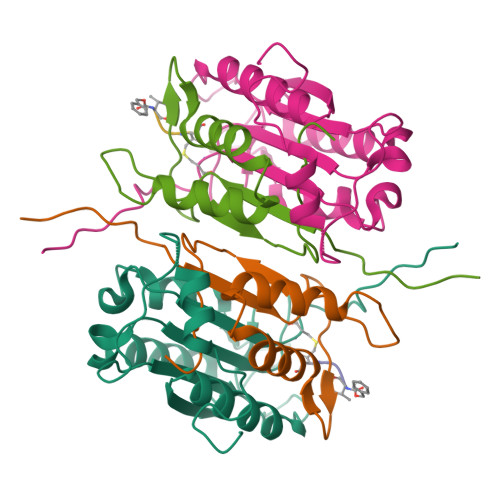
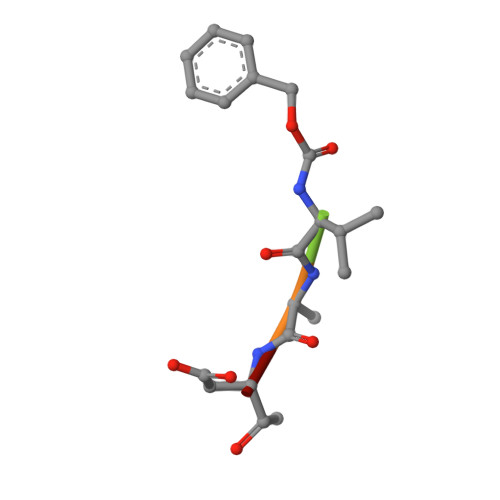
An allosteric circuit in caspase-1.
Datta, D., Scheer, J.M., Romanowski, M.J., Wells, J.A.(2008) J Mol Biology 381: 1157-1167
- PubMed: 18590738
- DOI: https://doi.org/10.1016/j.jmb.2008.06.040
- Primary Citation of Related Structures:
2H4W, 2H4Y, 2H51, 2H54 - PubMed Abstract:
Structural studies of caspase-1 reveal that the dimeric thiol protease can exist in two states: in an on-state, when the active site is occupied, or in an off-state, when the active site is empty or when the enzyme is bound by a synthetic allosteric ligand at the dimer interface approximately 15 A from the active site. A network of 21 hydrogen bonds from nine side chains connecting the active and allosteric sites change partners when going between the on-state and the off-state. Alanine-scanning mutagenesis of these nine side chains shows that only two of them-Arg286 and Glu390, which form a salt bridge-have major effects, causing 100- to 200-fold reductions in catalytic efficiency (k(cat)/K(m)). Two neighbors, Ser332 and Ser339, have minor effects, causing 4- to 7-fold reductions. A more detailed mutational analysis reveals that the enzyme is especially sensitive to substitutions of the salt bridge: even a homologous R286K substitution causes a 150-fold reduction in k(cat)/K(m). X-ray crystal structures of these variants suggest the importance of both the salt bridge interaction and the coordination of solvent water molecules near the allosteric binding pocket. Thus, only a small subset of side chains from the larger hydrogen bonding network is critical for activity. These form a contiguous set of interactions that run from one active site through the allosteric site at the dimer interface and onto the second active site. This subset constitutes a functional allosteric circuit or "hot wire" that promotes site-to-site coupling.
- Departments of Pharmaceutical Chemistry and Cellular and Molecular Pharmacology, University of California at San Francisco, San Francisco, CA 94143, USA.
Organizational Affiliation: