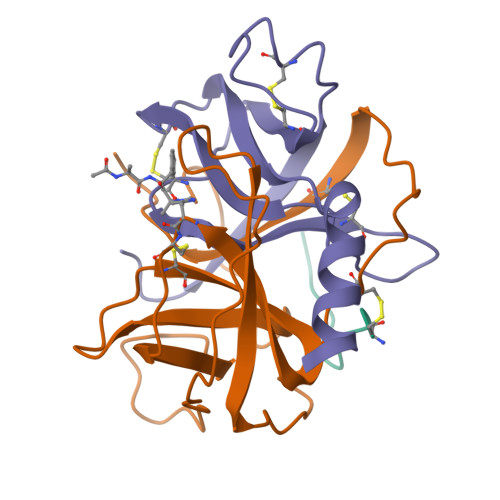
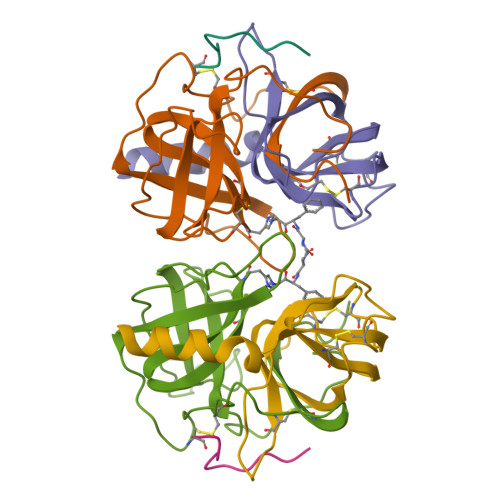
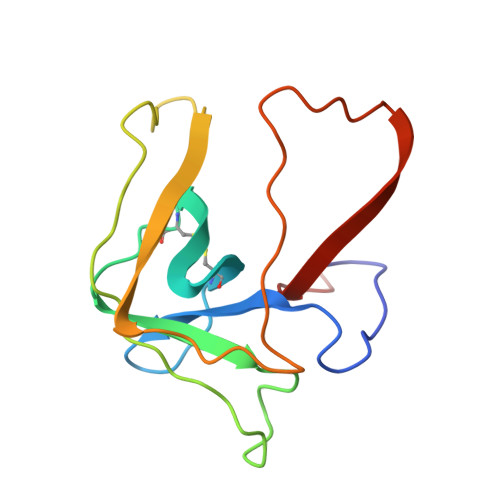
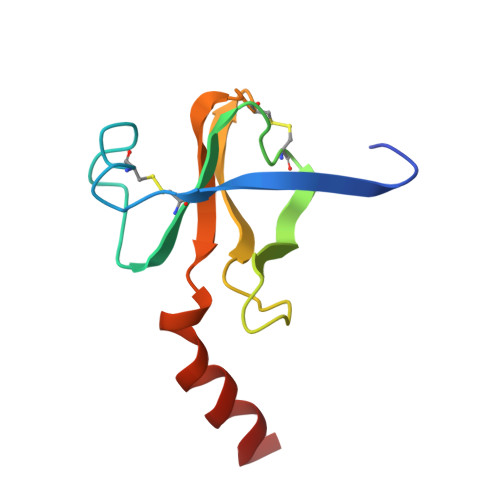
Three-dimensional structure of chymotrypsin inactivated with (2S)-N-acetyl-L-alanyl-L-phenylalanyl alpha-chloroethane: implications for the mechanism of inactivation of serine proteases by chloroketones.
Kreutter, K., Steinmetz, A.C., Liang, T.C., Prorok, M., Abeles, R.H., Ringe, D.(1994) Biochemistry 33: 13792-13800
- PubMed: 7947790
- DOI: https://doi.org/10.1021/bi00250a033
- Primary Citation of Related Structures:
2GMT - PubMed Abstract:
The reaction of enantiomerically pure (2S)-N-acetyl-L-alanyl-L-phenylalanyl alpha-chloroethane with gamma-chymotrypsin was studied as a probe of the mechanism of inactivation of serine proteases by peptidyl chloroalkanes. It was determined crystallographically that the peptidyl chloroethane alkylates His57 with retention of configuration at the chiral center, indicating a double displacement mechanism. We think it likely that a Ser195-epoxy ether adduct is an intermediate on the inactivation pathway, although other possibilities have not been disproven. Kinetic data reported by others [Angliker et al. (1988) Biochem. J. 256, 481-486] indicate that the epoxy ether intermediate is not an irreversibly inactivated form of enzyme [a conclusion confirmed experimentally (Prorok et al. (1994) Biochemistry 33, 9784-9790)] and that both ring closure of the tetrahedral intermediate to form the epoxy ether and ring opening by His57 partially limit the first-order rate constant for inactivation, ki. The peptidyl chloroethyl derivative adopts a very different active site conformation from that assumed by serine proteases inactivated by peptidyl chloromethanes. Positioning the chloroethyl derivative into the conformation adopted by chloromethyl derivatives would cause the extra methyl group to make a bad van der Waals contact with the inactivator P2 carbonyl carbon, thereby preventing the formation of the invariant hydrogen bond between the inactivator P1 amide nitrogen and the carbonyl group of Ser214. We conclude that the unusual conformation displayed by the chloroethyl derivative is caused by steric hindrance between the extra methyl group and the rest of the inactivator chain.
Organizational Affiliation:
Department of Biochemistry, Brandeis University, Waltham, Massachusetts 02254.