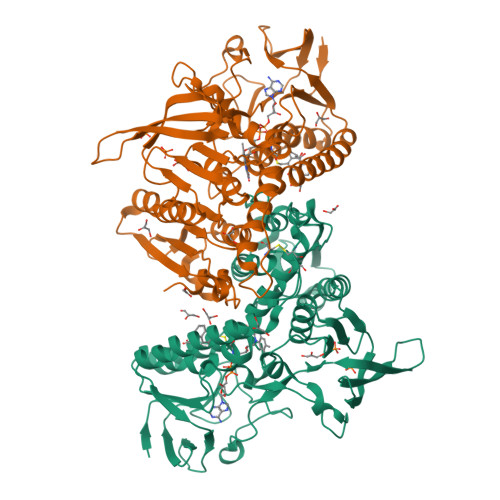
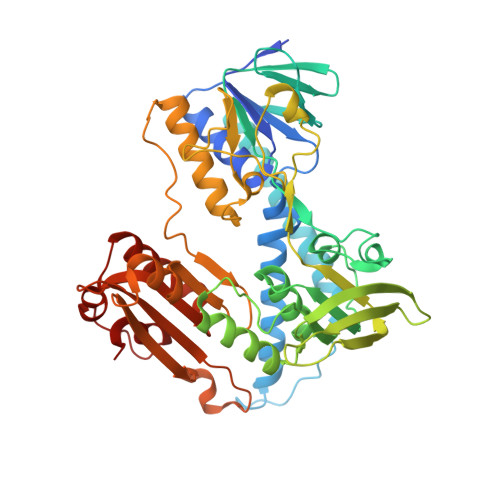
A fluoro analogue of the menadione derivative 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme
Bauer, H., Fritz-Wolf, K., Winzer, A., Little, S., Yardley, V., Vezin, H., Palfey, B., Schirmer, R.H., Davioud-Charvet, E.(2006) J Am Chem Soc 128: 10784-10794
- PubMed: 16910673
- DOI: https://doi.org/10.1021/ja061155v
- Primary Citation of Related Structures:
2GH5 - PubMed Abstract:
Glutathione reductase is an important housekeeping enzyme for redox homeostasis both in human cells and in the causative agent of tropical malaria, Plasmodium falciparum. Glutathione reductase inhibitors were shown to have anticancer and antimalarial activity per se and to contribute to the reversal of drug resistance. The development of menadione chemistry has led to the selection of 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid, called M(5), as a potent reversible and uncompetitive inhibitor of both human and P. falciparum glutathione reductases. Here we describe the synthesis and kinetic characterization of a fluoromethyl-M(5) analogue that acts as a mechanism-based inhibitor of both enzymes. In the course of enzymatic catalysis, the suicide substrate is activated by one- or two-electron reduction, and then a highly reactive quinone methide is generated upon elimination of the fluorine. Accordingly the human enzyme was found to be irreversibly inactivated with a k(inact) value of 0.4 +/- 0.2 min(-1). The crystal structure of the alkylated enzyme was solved at 1.7 A resolution. It showed the inhibitor to bind covalently to the active site Cys58 and to interact noncovalently with His467', Arg347, Arg37, and Tyr114. On the basis of the crystal structure of the inactivated human enzyme and stopped-flow kinetic studies with two- and four-electron-reduced forms of the unreacted P. falciparum enzyme, a mechanism is proposed which explains naphthoquinone reduction at the flavin of glutathione reductase.
Organizational Affiliation:
Biochemie-Zentrum der Universität Heidelberg, Im Neuenheimer Feld 504, D-69120 Heidelberg, Germany.