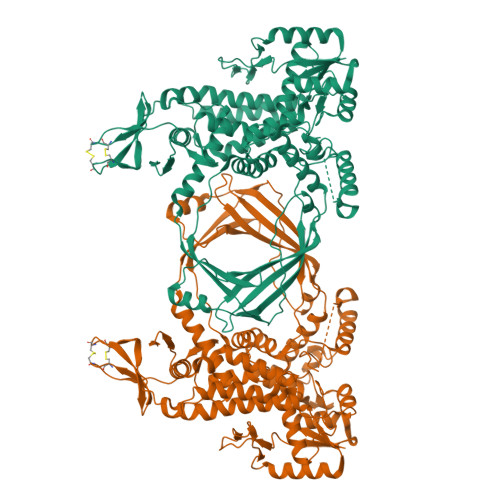
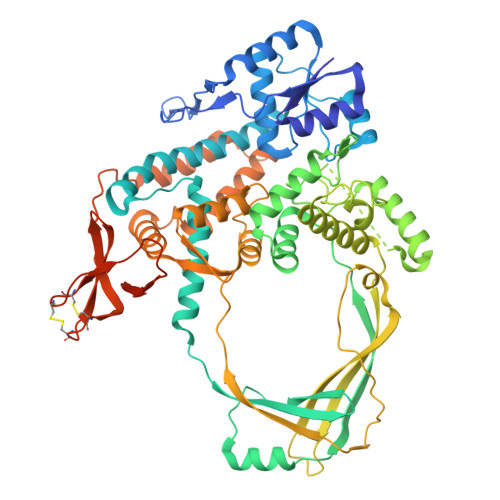
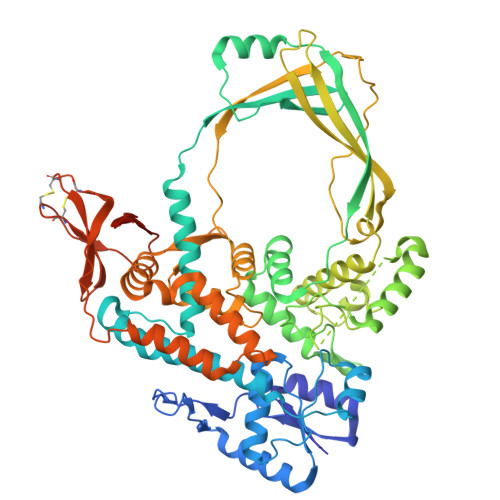
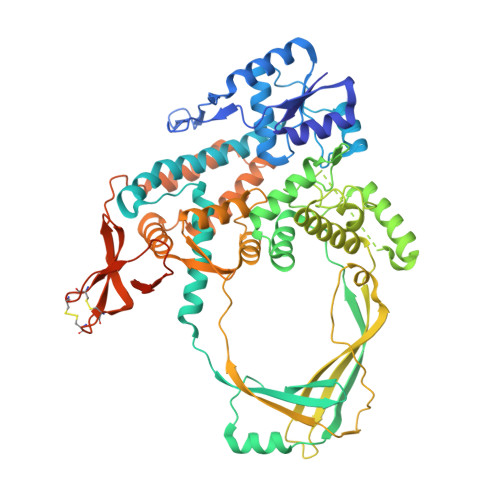
Crystal Structure of Full Length Topoisomerase I from Thermotoga maritima
Hansen, G., Harrenga, A., Wieland, B., Schomburg, D., Reinemer, P.(2006) J Mol Biology 358: 1328-1340
- PubMed: 16600296
- DOI: https://doi.org/10.1016/j.jmb.2006.03.012
- Primary Citation of Related Structures:
2GAI, 2GAJ - PubMed Abstract:
DNA topoisomerases are a family of enzymes altering the topology of DNA by concerted breakage and rejoining of the phosphodiester backbone of DNA. Bacterial and archeal type IA topoisomerases, including topoisomerase I, topoisomerase III, and reverse gyrase, are crucial in regulation of DNA supercoiling and maintenance of genetic stability. The crystal structure of full length topoisomerase I from Thermotoga maritima was determined at 1.7A resolution and represents an intact and fully active bacterial topoisomerase I. It reveals the torus-like structure of the conserved transesterification core domain comprising domains I-IV and a tightly associated C-terminal zinc ribbon domain (domain V) packing against domain IV of the core domain. The previously established zinc-independence of the functional activity of T.maritima topoisomerase I is further supported by its crystal structure as no zinc ion is bound to domain V. However, the structural integrity is preserved by the formation of two disulfide bridges between the four Zn-binding cysteine residues. A functional role of domain V in DNA binding and recognition is suggested and discussed in the light of the structure and previous biochemical findings. In addition, implications for bacterial topoisomerases I are provided.
Organizational Affiliation:
Bayer HealthCare AG, Pharma R and D Europe, Enabling Technologies, D-42096 Wuppertal, Germany.