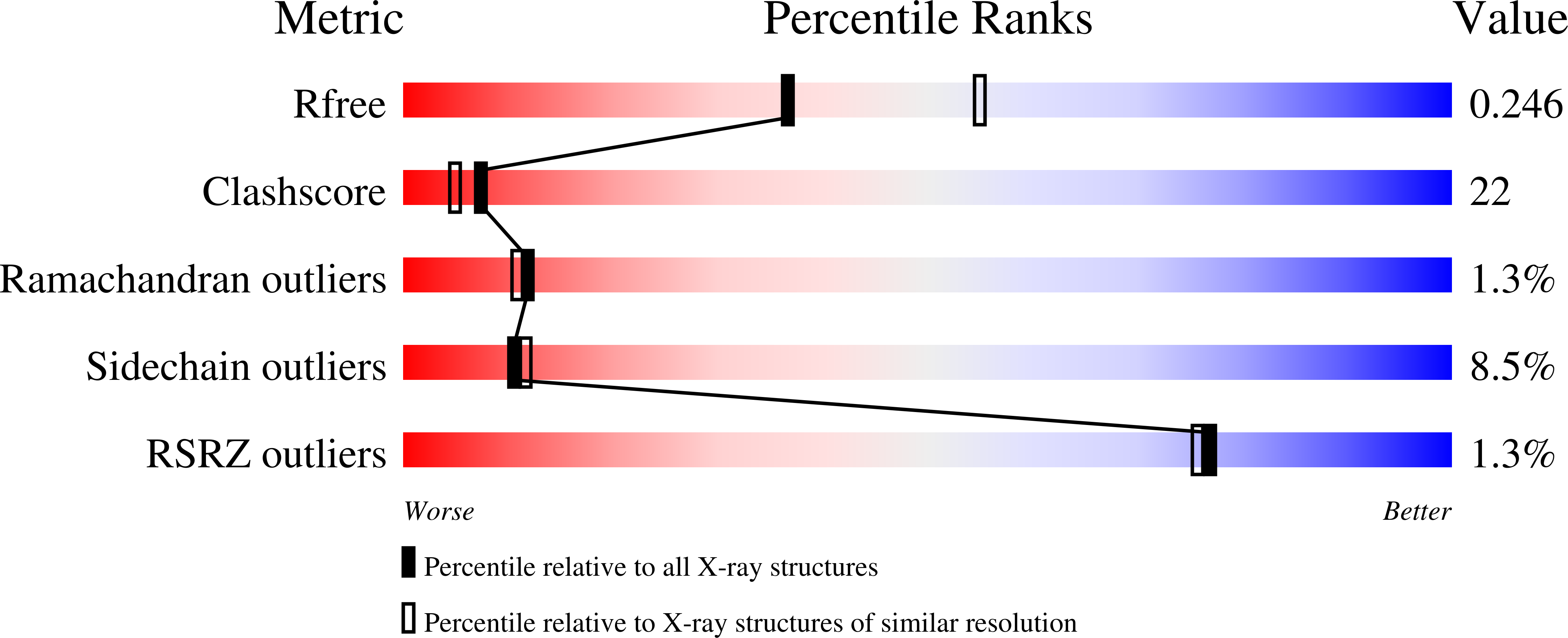
Structure of the calmodulin alphaII-spectrin complex provides insight into the regulation of cell plasticity.
Simonovic, M., Zhang, Z., Cianci, C.D., Steitz, T.A., Morrow, J.S.(2006) J Biol Chem 281: 34333-34340
- PubMed: 16945920
- DOI: https://doi.org/10.1074/jbc.M604613200
- Primary Citation of Related Structures:
2FOT - PubMed Abstract:
AlphaII-spectrin is a major cortical cytoskeletal protein contributing to membrane organization and integrity. The Ca2+-activated binding of calmodulin to an unstructured insert in the 11th repeat unit of alphaII-spectrin enhances the susceptibility of spectrin to calpain cleavage but abolishes its sensitivity to several caspases and to at least one bacterially derived pathologic protease. Other regulatory inputs including phosphorylation by c-Src also modulate the proteolytic susceptibility of alphaII-spectrin. These pathways, acting through spectrin, appear to control membrane plasticity and integrity in several cell types. To provide a structural basis for understanding these crucial biological events, we have solved the crystal structure of a complex between bovine calmodulin and the calmodulin-binding domain of human alphaII-spectrin (Protein Data Bank ID code 2FOT). The structure revealed that the entire calmodulin-spectrin-binding interface is hydrophobic in nature. The spectrin domain is also unique in folding into an amphiphilic helix once positioned within the calmodulin-binding groove. The structure of this complex provides insight into the mechanisms by which calmodulin, calpain, caspase, and tyrosine phosphorylation act on spectrin to regulate essential cellular processes.
Organizational Affiliation:
Howard Hughes Medical Institute, Yale University, New Haven, Connecticut 06520, USA.