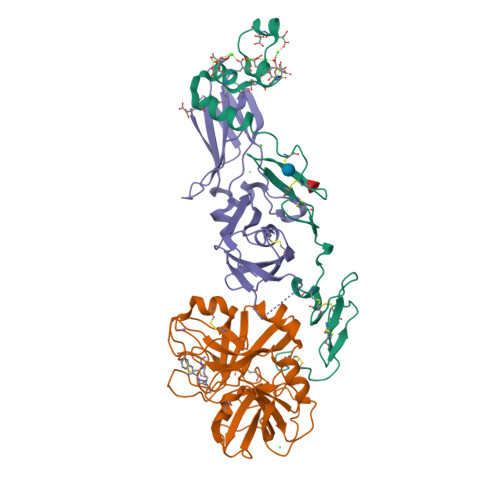
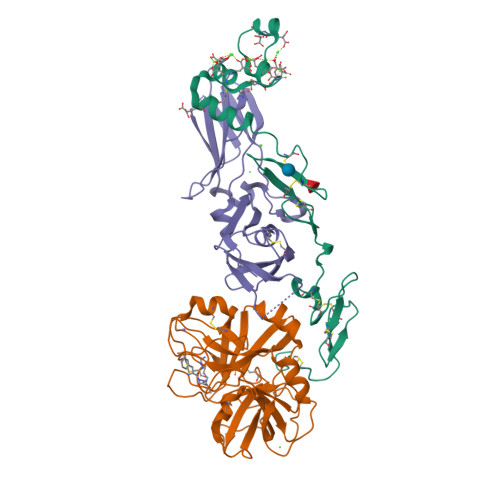
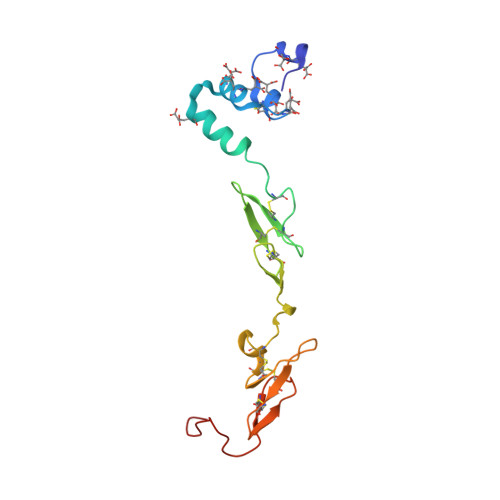
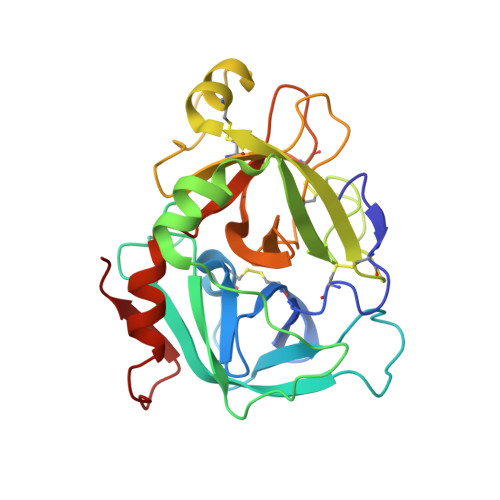
High Resolution Structures of p-Aminobenzamidine- and Benzamidine-VIIa/Soluble Tissue Factor: Unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+ and Zn2+ sites in factor VIIa
Bajaj, S.P., Schmidt, A.E., Agah, S., Bajaj, M.S., Padmanabhan, K.(2006) J Biological Chem 281: 24873-24888
- PubMed: 16757484
- DOI: https://doi.org/10.1074/jbc.M509971200
- Primary Citation of Related Structures:
2A2Q, 2AER, 2FIR - PubMed Abstract:
Factor VIIa (FVIIa) consists of a gamma-carboxyglutamic acid (Gla) domain, two epidermal growth factor-like domains, and a protease domain. FVIIa binds seven Ca(2+) ions in the Gla, one in the EGF1, and one in the protease domain. However, blood contains both Ca(2+) and Mg(2+), and the Ca(2+) sites in FVIIa that could be specifically occupied by Mg(2+) are unknown. Furthermore, FVIIa contains a Na(+) and two Zn(2+) sites, but ligands for these cations are undefined. We obtained p-aminobenzamidine-VIIa/soluble tissue factor (sTF) crystals under conditions containing Ca(2+), Mg(2+), Na(+), and Zn(2+). The crystal diffracted to 1.8A resolution, and the final structure has an R-factor of 19.8%. In this structure, the Gla domain has four Ca(2+) and three bound Mg(2+). The EGF1 domain contains one Ca(2+) site, and the protease domain contains one Ca(2+), one Na(+), and two Zn(2+) sites. (45)Ca(2+) binding in the presence/absence of Mg(2+) to FVIIa, Gla-domainless FVIIa, and prothrombin fragment 1 supports the crystal data. Furthermore, unlike in other serine proteases, the amide N of Gly(193) in FVIIa points away from the oxyanion hole in this structure. Importantly, the oxyanion hole is also absent in the benzamidine-FVIIa/sTF structure at 1.87A resolution. However, soaking benzamidine-FVIIa/sTF crystals with d-Phe-Pro-Arg-chloromethyl ketone results in benzamidine displacement, d-Phe-Pro-Arg incorporation, and oxyanion hole formation by a flip of the 192-193 peptide bond in FVIIa. Thus, it is the substrate and not the TF binding that induces oxyanion hole formation and functional active site geometry in FVIIa. Absence of oxyanion hole is unusual and has biologic implications for FVIIa macromolecular substrate specificity and catalysis.
Organizational Affiliation:
Protein Science Laboratory, UCLA/Orthopaedic Hospital, Department of Orthopaedic Surgery and Molecular Biology Institute, UCLA, Los Angeles, California 90095, USA.