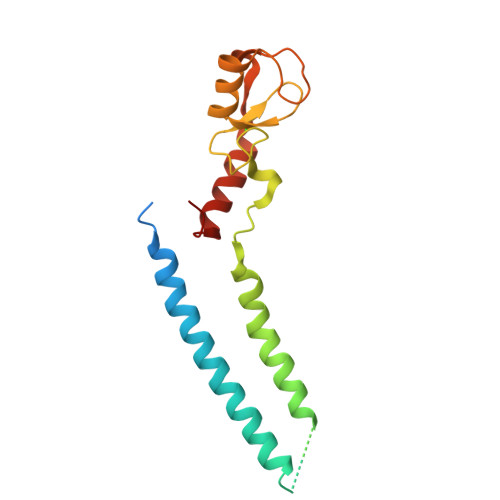
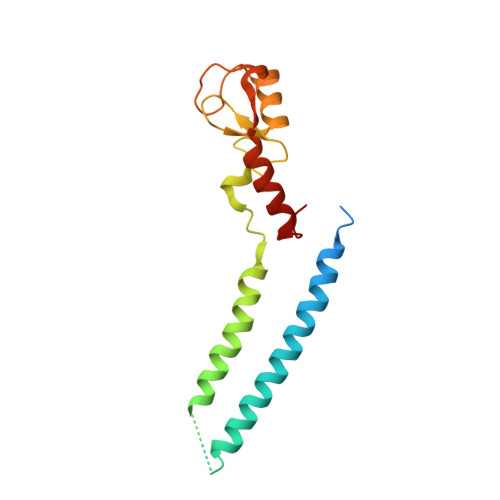
Structure and Interactions of the Helical and U-Box Domains of CHIP, the C Terminus of HSP70 Interacting Protein.
Xu, Z., Devlin, K.I., Ford, M.G., Nix, J.C., Qin, J., Misra, S.(2006) Biochemistry 45: 4749-4759
- PubMed: 16605243
- DOI: https://doi.org/10.1021/bi0601508
- Primary Citation of Related Structures:
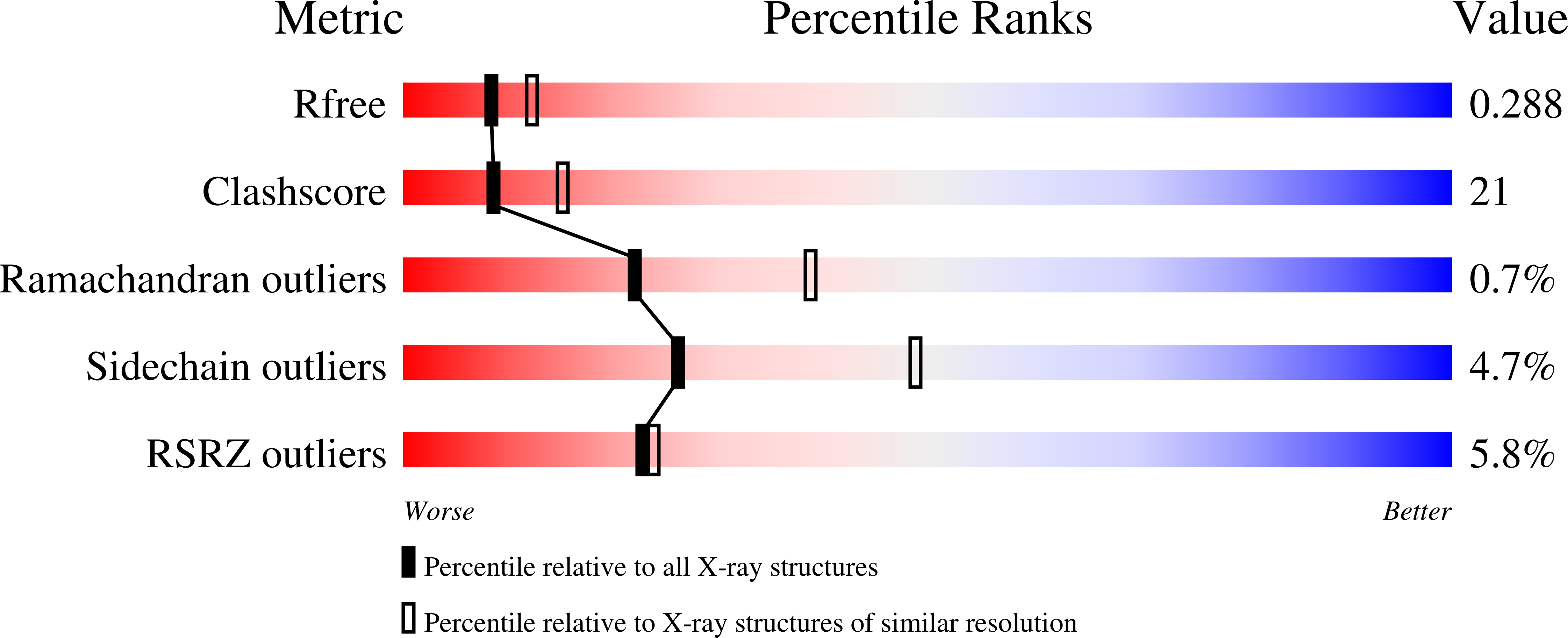
2F42 - PubMed Abstract:
The heat-shock proteins Hsp70 and Hsp90 play a crucial role in regulating protein quality control both by refolding and by preventing the aggregation of misfolded proteins. It has recently been shown that Hsp70 and Hsp90 act not only in protein refolding but also cooperate with the C terminus of Hsp70 interacting protein (CHIP), a multidomain ubiquitin ligase, to mediate the degradation of unfolded proteins. We present the crystal structure of the helical linker domain and U-box domain of zebrafish CHIP (DrCHIP-HU). The structure of DrCHIP-HU shows a symmetric homodimer. The conformation of the helical linker domains and the relative positions of the helical and U-box domains differ substantially in DrCHIP-HU from those in a recently published structure of an asymmetric dimer of mammalian (mouse) CHIP. We used an in vitro ubiquitination assay to identify residues, located on two long loops and a central alpha helix of the CHIP U-box domain, that are important for interacting with the ubiquitin-conjugating enzyme UbcH5b. In addition, we used NMR spectroscopy to define a complementary interaction surface located on the N-terminal alpha helix and the L4 and L7 loops of UbcH5b. Our results provide insights into conformational variability in the domain arrangement of CHIP and into U-box-mediated recruitment of UbcH5b for the ubiquitination of Hsp70 and Hsp90 substrates.
Organizational Affiliation:
Department of Molecular Cardiology, The Cleveland Clinic Foundation, Cleveland, Ohio 44195, USA.