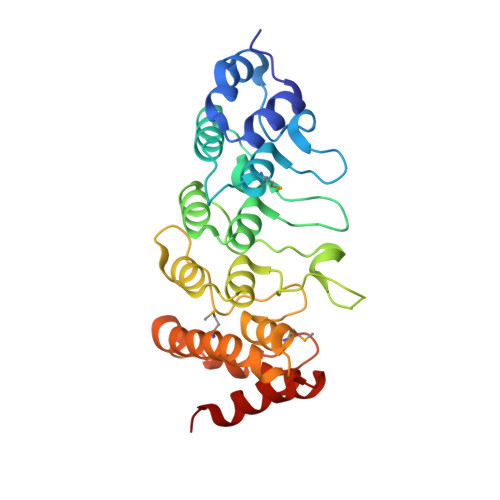
Crystal structure of the human TRPV2 channel ankyrin repeat domain.
McCleverty, C.J., Koesema, E., Patapoutian, A., Lesley, S.A., Kreusch, A.(2006) Protein Sci 15: 2201-2206
- PubMed: 16882997
- DOI: https://doi.org/10.1110/ps.062357206
- Primary Citation of Related Structures:
2F37 - PubMed Abstract:
TRPV channels are important polymodal integrators of noxious stimuli mediating thermosensation and nociception. An ankyrin repeat domain (ARD), which is a common protein-protein recognition domain, is conserved in the N-terminal intracellular domain of all TRPV channels and predicted to contain three to four ankyrin repeats. Here we report the first structure from the TRPV channel subfamily, a 1.7 A resolution crystal structure of the human TRPV2 ARD. Our crystal structure reveals a six ankyrin repeat stack with multiple insertions in each repeat generating several unique features compared with a canonical ARD. The surface typically used for ligand recognition, the ankyrin groove, contains extended loops with an exposed hydrophobic patch and a prominent kink resulting from a large rotational shift of the last two repeats. The TRPV2 ARD provides the first structural insight into a domain that coordinates nociceptive sensory transduction and is likely to be a prototype for other TRPV channel ARDs.
Organizational Affiliation:
Genomics Institute of the Novartis Research Foundation, CA 92121, USA.